Pigment epithelium-derived factor (PEDF) promotes tumor cell death by inducing macrophage membrane tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)
- PMID: 21846721
- PMCID: PMC3195575
- DOI: 10.1074/jbc.M111.266064
Pigment epithelium-derived factor (PEDF) promotes tumor cell death by inducing macrophage membrane tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)
Abstract
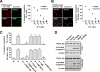
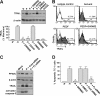
Pigment epithelium-derived factor (PEDF) is an intrinsic anti-angiogenic factor and a potential anti-tumor agent. The tumoricidal mechanism of PEDF, however, has not been fully elucidated. Here we report that PEDF induces the apoptosis of TC-1 and SK-Hep-1 tumor cells when they are cocultured with bone marrow-derived macrophages (BMDMs). This macrophage-mediated tumor killing is prevented by blockage of TNF-related apoptosis-inducing ligand (TRAIL) following treatment with the soluble TRAIL receptor. PEDF also increases the amount of membrane-bound TRAIL on cultured mouse BMDMs and on macrophages surrounding subcutaneous tumors. PEDF-induced tumor killing and TRAIL induction are abrogated by peroxisome proliferator-activated receptor γ (PPARγ) antagonists or small interfering RNAs targeting PPARγ. PEDF also induces PPARγ in BMDMs. Furthermore, the activity of the TRAIL promoter in human macrophages is increased by PEDF stimulation. Chromatin immunoprecipitation and DNA pull-down assays confirmed that endogenous PPARγ binds to a functional PPAR-response element (PPRE) in the TRAIL promoter, and mutation of this PPRE abolishes the binding of the PPARγ-RXRα heterodimer. Also, PPARγ-dependent transactivation and PPARγ-RXRα binding to this PPRE are prevented by PPARγ antagonists. Our results provide a novel mechanism for the tumoricidal activity of PEDF, which involves tumor cell killing via PPARγ-mediated TRAIL induction in macrophages.
Figures


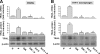
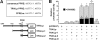

Similar articles
-
Pigment epithelium-derived factor induces THP-1 macrophage apoptosis and necrosis by the induction of the peroxisome proliferator-activated receptor gamma.Mol Immunol. 2008 Feb;45(4):898-909. doi: 10.1016/j.molimm.2007.08.004. Epub 2007 Sep 17. Mol Immunol. 2008. PMID: 17870167
-
Proapoptotic PEDF functional peptides inhibit prostate tumor growth--a mechanistic study.Biochem Pharmacol. 2014 Dec 1;92(3):425-37. doi: 10.1016/j.bcp.2014.09.012. Epub 2014 Sep 28. Biochem Pharmacol. 2014. PMID: 25261795
-
Pigment epithelium-derived factor induces interleukin-10 expression in human macrophages by induction of PPAR gamma.Life Sci. 2010 Jul 3;87(1-2):26-35. doi: 10.1016/j.lfs.2010.05.007. Epub 2010 May 27. Life Sci. 2010. PMID: 20553736
-
Cancer cell apoptotic pathways mediated by PEDF: prospects for therapy.Trends Mol Med. 2009 Oct;15(10):461-7. doi: 10.1016/j.molmed.2009.08.003. Epub 2009 Sep 25. Trends Mol Med. 2009. PMID: 19783213 Review.
-
The role of PEDF in tumor growth and metastasis.Curr Mol Med. 2010 Apr;10(3):292-5. doi: 10.2174/156652410791065327. Curr Mol Med. 2010. PMID: 20236054 Review.
Cited by
-
In vitro heterochronic parabiosis identifies pigment epithelium-derived factor as a systemic mediator of rejuvenation by young blood.bioRxiv [Preprint]. 2024 May 5:2024.05.02.592258. doi: 10.1101/2024.05.02.592258. bioRxiv. 2024. PMID: 38746475 Free PMC article. Preprint.
-
Melanoma Cells Block PEDF Production in Fibroblasts to Induce the Tumor-Promoting Phenotype of Cancer-Associated Fibroblasts.Cancer Res. 2016 Apr 15;76(8):2265-76. doi: 10.1158/0008-5472.CAN-15-2468. Epub 2016 Feb 26. Cancer Res. 2016. PMID: 26921338 Free PMC article.
-
Eye drop delivery of pigment epithelium-derived factor-34 promotes retinal ganglion cell neuroprotection and axon regeneration.Mol Cell Neurosci. 2015 Sep;68:212-21. doi: 10.1016/j.mcn.2015.08.001. Epub 2015 Aug 8. Mol Cell Neurosci. 2015. PMID: 26260110 Free PMC article.
-
The Role of TRAIL/DRs in the Modulation of Immune Cells and Responses.Cancers (Basel). 2019 Sep 30;11(10):1469. doi: 10.3390/cancers11101469. Cancers (Basel). 2019. PMID: 31574961 Free PMC article. Review.
-
Pigment epithelium-derived factor: clinical significance in estrogen-dependent tissues and its potential in cancer therapy.Iran J Basic Med Sci. 2015 Sep;18(9):837-55. Iran J Basic Med Sci. 2015. PMID: 26523216 Free PMC article. Review.
References
-
- Dawson D. W., Volpert O. V., Gillis P., Crawford S. E., Xu H., Benedict W., Bouck N. P. (1999) Science 285, 245–248 - PubMed
-
- Zhang S. X., Wang J. J., Gao G., Shao C., Mott R., Ma J. X. (2006) FASEB J. 20, 323–335 - PubMed
-
- Tombran-Tink J., Mazuruk K., Rodriguez I. R., Chung D., Linker T., Englander E., Chader G. J. (1996) Mol. Vis. 2, 11. - PubMed
-
- Guan M., Pang C. P., Yam H. F., Cheung K. F., Liu W. W., Lu Y. (2004) Cancer Gene Ther. 11, 325–332 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

