Insights into folate/FAD-dependent tRNA methyltransferase mechanism: role of two highly conserved cysteines in catalysis
- PMID: 21846722
- PMCID: PMC3196131
- DOI: 10.1074/jbc.M111.256966
Insights into folate/FAD-dependent tRNA methyltransferase mechanism: role of two highly conserved cysteines in catalysis
Abstract
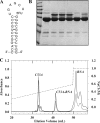
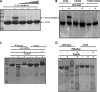
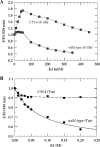
The flavoprotein TrmFO methylates specifically the C5 carbon of the highly conserved uridine 54 in tRNAs. Contrary to most methyltransferases, the 1-carbon unit transferred by TrmFO derives from 5,10-methylenetetrahydrofolate and not from S-adenosyl-L-methionine. The enzyme also employs the FAD hydroquinone as a reducing agent of the C5 methylene U54-tRNA intermediate in vitro. By analogy with the catalytic mechanism of thymidylate synthase ThyA, a conserved cysteine located near the FAD isoalloxazine ring was proposed to act as a nucleophile during catalysis. Here, we mutated this residue (Cys-53 in Bacillus subtilis TrmFO) to alanine and investigated its functional role. Biophysical characterization of this variant demonstrated the major structural role of Cys-53 in maintaining both the integrity and plasticity of the flavin binding site. Unexpectedly, gel mobility shift assays showed that, like the wild-type enzyme, the inactive C53A variant was capable of forming a covalent complex with a 5-fluorouridine-containing mini-RNA. This result confirms the existence of a covalent intermediate during catalysis but rules out a nucleophilic role for Cys-53. To identify the actual nucleophile, two other strictly conserved cysteines (Cys-192 and Cys-226) that are relatively far from the active site were replaced with alanine, and a double mutant C53A/C226A was generated. Interestingly, only mutations that target Cys-226 impeded TrmFO from forming a covalent complex and methylating tRNA. Altogether, we propose a revised mechanism for the m(5)U54 modification catalyzed by TrmFO, where Cys-226 attacks the C6 atom of the uridine, and Cys-53 plays the role of the general base abstracting the C5 proton.
Figures



Similar articles
-
A catalytic intermediate and several flavin redox states stabilized by folate-dependent tRNA methyltransferase from Bacillus subtilis.Biochemistry. 2011 Jun 14;50(23):5208-19. doi: 10.1021/bi1019463. Epub 2011 May 18. Biochemistry. 2011. PMID: 21561081
-
The tRNA recognition mechanism of folate/FAD-dependent tRNA methyltransferase (TrmFO).J Biol Chem. 2012 Dec 14;287(51):42480-94. doi: 10.1074/jbc.M112.390112. Epub 2012 Oct 24. J Biol Chem. 2012. PMID: 23095745 Free PMC article.
-
Activation of a unique flavin-dependent tRNA-methylating agent.Biochemistry. 2013 Dec 10;52(49):8949-56. doi: 10.1021/bi4013879. Epub 2013 Nov 20. Biochemistry. 2013. PMID: 24228791
-
Flavin-Dependent Methylation of RNAs: Complex Chemistry for a Simple Modification.J Mol Biol. 2016 Dec 4;428(24 Pt B):4867-4881. doi: 10.1016/j.jmb.2016.10.031. Epub 2016 Nov 5. J Mol Biol. 2016. PMID: 27825927 Review.
-
In vitro detection of the enzymatic activity of folate-dependent tRNA (Uracil-54,-C5)-methyltransferase: evolutionary implications.Methods Enzymol. 2007;425:103-19. doi: 10.1016/S0076-6879(07)25004-9. Methods Enzymol. 2007. PMID: 17673080 Review.
Cited by
-
Structural and functional insights into tRNA binding and adenosine N1-methylation by an archaeal Trm10 homologue.Nucleic Acids Res. 2016 Jan 29;44(2):940-53. doi: 10.1093/nar/gkv1369. Epub 2015 Dec 15. Nucleic Acids Res. 2016. PMID: 26673726 Free PMC article.
-
Methylated nucleosides in tRNA and tRNA methyltransferases.Front Genet. 2014 May 23;5:144. doi: 10.3389/fgene.2014.00144. eCollection 2014. Front Genet. 2014. PMID: 24904644 Free PMC article. Review.
-
Diversity in mechanism and function of tRNA methyltransferases.RNA Biol. 2015;12(4):398-411. doi: 10.1080/15476286.2015.1008358. RNA Biol. 2015. PMID: 25626150 Free PMC article. Review.
-
Mechanisms and inhibition of uracil methylating enzymes.Bioorg Chem. 2012 Aug;43:37-43. doi: 10.1016/j.bioorg.2011.11.005. Epub 2011 Nov 27. Bioorg Chem. 2012. PMID: 22172597 Free PMC article. Review.
-
Structural and functional analyses of the archaeal tRNA m2G/m22G10 methyltransferase aTrm11 provide mechanistic insights into site specificity of a tRNA methyltransferase that contains common RNA-binding modules.Nucleic Acids Res. 2016 Jul 27;44(13):6377-90. doi: 10.1093/nar/gkw561. Epub 2016 Jun 20. Nucleic Acids Res. 2016. PMID: 27325738 Free PMC article.
References
-
- Delk A. S., Nagle D. P., Jr., Rabinowitz J. C. (1980) J. Biol. Chem. 255, 4387–4390 - PubMed
-
- Hamdane D., Guérineau V., Un S., Golinelli-Pimpaneau B. (2011) Biochemistry 50, 5208–5219 - PubMed
-
- Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. (1991) Biochemistry 30, 11018–11025 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

