The downstream atpE cistron is efficiently translated via its own cis-element in partially overlapping atpB-atpE dicistronic mRNAs in chloroplasts
- PMID: 21846772
- PMCID: PMC3241655
- DOI: 10.1093/nar/gkr644
The downstream atpE cistron is efficiently translated via its own cis-element in partially overlapping atpB-atpE dicistronic mRNAs in chloroplasts
Abstract
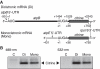
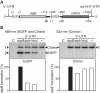
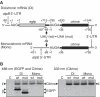
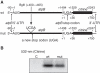
The chloroplast atpB and atpE genes encode subunits β and ε of the ATP synthase, respectively. They are co-transcribed as dicistronic mRNAs in flowering plants. An unusual feature is an overlap (AUGA) of the atpB stop codon (UGA) with the atpE start codon (AUG). Hence, atpE translation has been believed to depend on atpB translation (i.e. translational coupling). Using an in vitro translation system from tobacco chloroplasts, we showed that both atpB and atpE cistrons are translated from the tobacco dicistronic mRNA, and that the efficiency of atpB translation is higher than that of atpE translation. When the atpB 5'-UTR was replaced with lower efficiency 5'-UTRs, atpE translation was higher than atpB translation. Removal of the entire atpB 5'-UTR arrested atpB translation but atpE translation still proceeded. Introduction of a premature stop codon in the atpB cistron did not abolish atpE translation. These results indicate that atpE translation is independent of atpB translation. Mutation analysis showed that the atpE cistron possesses its own cis-element(s) for translation, located ~25 nt upstream from the start codon.
Figures


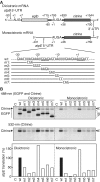
References
-
- Sugiura M. The chloroplast genome. Plant Mol. Biol. 1992;19:149–168. - PubMed
-
- Bock R. Structure, function, and inheritance of plastid genomes. In: Bock R, editor. Cell and Molecular Biology of Plastids. Vol. 19. Potsdam-Golm: Springer; 2007. pp. 29–63.
-
- Deng XW, Gruissem W. Control of plastid gene expression during development: the limited role of transcriptional regulation. Cell. 1987;49:379–387. - PubMed
-
- Mullet JE. Chloroplast development and gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988;39:475–502.
-
- Marín-Navarro J, Manuell AL, Wu J, Mayfield SP. Chloroplast translation regulation. Photosynth. Res. 2007;94:359–374. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

