PIKE-mediated PI3-kinase activity is required for AMPA receptor surface expression
- PMID: 21847098
- PMCID: PMC3199380
- DOI: 10.1038/emboj.2011.281
PIKE-mediated PI3-kinase activity is required for AMPA receptor surface expression
Abstract
AMPAR (α-amino-3-hydroxy-5-methyl-isoxazole-4-propionic acid receptor) is an ion channel involved in the formation of synaptic plasticity. However, the molecular mechanism that couples plasticity stimuli to the trafficking of postsynaptic AMPAR remains poorly understood. Here, we show that PIKE (phosphoinositide 3-kinase enhancer) GTPases regulate neuronal AMPAR activity by promoting GluA2/GRIP1 association. PIKE-L directly interacts with both GluA2 and GRIP1 and forms a tertiary complex upon glycine-induced NMDA receptor activation. PIKE-L is also essential for glycine-induced GluA2-associated PI3K activation. Genetic ablation of PIKE (PIKE(-/-)) in neurons suppresses GluA2-associated PI3K activation, therefore inhibiting the subsequent surface expression of GluA2 and the formation of long-term potentiation. Our findings suggest that PIKE-L is a critical factor in controlling synaptic AMPAR insertion.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

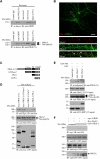
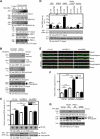
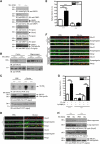
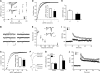
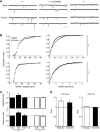
References
-
- Ahn JY, Rong R, Kroll TG, Van Meir EG, Snyder SH, Ye K (2004) PIKE (phosphatidylinositol 3-kinase enhancer)-A GTPase stimulates Akt activity and mediates cellular invasion. J Biol Chem 279: 16441–16451 - PubMed
-
- Bliss TV, Collingridge GL (1993) A synaptic model of memory: long-term potentiation in the hippocampus. Nature 361: 31–39 - PubMed
-
- Burnashev N, Monyer H, Seeburg PH, Sakmann B (1992) Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron 8: 189–198 - PubMed
-
- Capocchi G, Zampolini M, Larson J (1992) Theta burst stimulation is optimal for induction of LTP at both apical and basal dendritic synapses on hippocampal CA1 neurons. Brain Res 591: 332–336 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

