Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen
- PMID: 21847113
- PMCID: PMC3202794
- DOI: 10.1038/msb.2011.55
Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen
Abstract
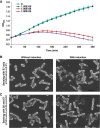
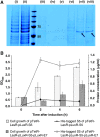
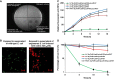
Synthetic biology aims to systematically design and construct novel biological systems that address energy, environment, and health issues. Herein, we describe the development of a synthetic genetic system, which comprises quorum sensing, killing, and lysing devices, that enables Escherichia coli to sense and kill a pathogenic Pseudomonas aeruginosa strain through the production and release of pyocin. The sensing, killing, and lysing devices were characterized to elucidate their detection, antimicrobial and pyocin release functionalities, which subsequently aided in the construction of the final system and the verification of its designed behavior. We demonstrated that our engineered E. coli sensed and killed planktonic P. aeruginosa, evidenced by 99% reduction in the viable cells. Moreover, we showed that our engineered E. coli inhibited the formation of P. aeruginosa biofilm by close to 90%, leading to much sparser and thinner biofilm matrices. These results suggest that E. coli carrying our synthetic genetic system may provide a novel synthetic biology-driven antimicrobial strategy that could potentially be applied to fighting P. aeruginosa and other infectious pathogens.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Comment in
-
Synthetic biology: licensing bacteria to kill.Nat Rev Microbiol. 2011 Sep 12;9(10):699. doi: 10.1038/nrmicro2660. Nat Rev Microbiol. 2011. PMID: 21909129 No abstract available.
-
Licensing bacteria to kill.Nat Rev Genet. 2011 Sep 13;12(10):668. doi: 10.1038/nrg3075. Nat Rev Genet. 2011. PMID: 21915106 No abstract available.
References
-
- Anderson JC, Clarke EJ, Arkin AP, Voigt CA (2006) Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol 355: 619–627 - PubMed
-
- Baba T, Schneewind O (1998) Instruments of microbial warfare: bacteriocin synthesis, toxicity and immunity. Trends Microbiol 6: 66–71 - PubMed
-
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 - PubMed
-
- Brook I (2005) The role of bacterial interference in otitis, sinusitis and tonsillitis. Otolaryngol Head Neck Surg 133: 139–146 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

