Mass spectrometry-based protein footprinting characterizes the structures of oligomeric apolipoprotein E2, E3, and E4
- PMID: 21848287
- PMCID: PMC3177987
- DOI: 10.1021/bi200911c
Mass spectrometry-based protein footprinting characterizes the structures of oligomeric apolipoprotein E2, E3, and E4
Abstract
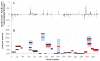
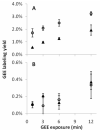
The three common isoforms of apolipoprotein E (ApoE) differ at two sites in their 299 amino acid sequence; these differences modulate the structure of ApoE to affect profoundly the isoform associations with disease. The ε4 allele in particular is strongly associated with Alzheimer's disease. The study of the structural effects of these mutation sites in aqueous media is hampered by the aggregation proclivity of each ApoE isoform. Hence, understanding the differences between isoforms has thus far relied on lower resolution biophysical measurements, mutagenesis, homology studies, and the use of truncated ApoE variants. In this study, we report two comparative studies of the ApoE family by using the mass spectrometry-based protein footprinting methods of FPOP and glycine ethyl ester (GEE) labeling. The first experiment examines the three full-length WT isoforms in their tetrameric state and finds that the overall structures are similar, with the exception of M108 in ApoE4 which is more solvent-accessible in this isoform than in ApoE2 and ApoE3. The second experiment provides clear evidence, from a comparison of the footprinting results of the wild-type proteins and a monomeric mutant, that several residues in regions 183-205 and 232-251 are involved in self-association.
Figures


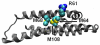

References
-
- Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. - PubMed
-
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. - PubMed
-
- Weisgraber KH. Advances in Protein Chemistry. Academic Press; 1994. Apolipoprotein E: Structure-Function Relationships; pp. 249–302. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

