HIV type 1 Gag as a target for antiviral therapy
- PMID: 21848364
- PMCID: PMC3251841
- DOI: 10.1089/AID.2011.0230
HIV type 1 Gag as a target for antiviral therapy
Abstract
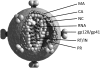
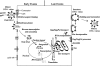
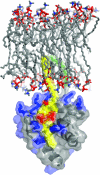
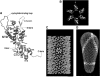
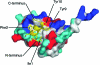
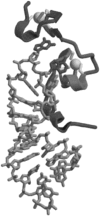
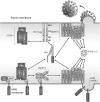
The Gag proteins of HIV-1 are central players in virus particle assembly, release, and maturation, and also function in the establishment of a productive infection. Despite their importance throughout the replication cycle, there are currently no approved antiretroviral therapies that target the Gag precursor protein or any of the mature Gag proteins. Recent progress in understanding the structural and cell biology of HIV-1 Gag function has revealed a number of potential Gag-related targets for possible therapeutic intervention. In this review, we summarize our current understanding of HIV-1 Gag and suggest some approaches for the development of novel antiretroviral agents that target Gag.
Figures
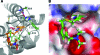

References
-
- Berger EA. Murphy PM. Farber JM. Chemokine receptors as HIV‐1 coreceptors: Roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. - PubMed
-
- Doms RW. Beyond receptor expression: The influence of receptor conformation, density, and affinity in HIV-1 infection. Virology. 2000;276(2):229–237. - PubMed
-
- Freed EO. Martin MA. HIVs, their replication. In: Knipe DM, editor; Howley PM, editor. Fields Virology. Lippincott, Williams & Wilkins; Philadelphia: 2007. pp. 2107–2185.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

