Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production
- PMID: 21849504
- PMCID: PMC3190816
- DOI: 10.1074/jbc.M111.268649
Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production
Abstract
In all forms of life, rRNAs for the small and large ribosomal subunit are co-transcribed as a single transcript. Although this ensures the equimolar production of rRNAs, it requires the endonucleolytic separation of pre-rRNAs to initiate rRNA production. In yeast, processing of the primary transcript encoding 18 S, 5.8 S, and 25 S rRNAs has been studied extensively. Nevertheless, most nucleases remain to be identified. Here, we show that Rcl1, conserved in all eukaryotes, cleaves pre-rRNA at so-called site A(2), a co-transcriptional cleavage step that separates rRNAs destined for the small and large subunit. Recombinant Rcl1 cleaves pre-rRNA mimics at site A(2) in a reaction that is sensitive to nearby RNA mutations that inhibit cleavage in vivo. Furthermore, mutations in Rcl1 disrupt rRNA processing at site A(2) in vivo and in vitro. Together, these results demonstrate that the role of Rcl1 in eukaryotic pre-rRNA processing is identical to that of RNase III in bacteria: to co-transcriptionally separate the pre-rRNAs destined for the small and large subunit. Furthermore, because Rcl1 has no homology to other known endonucleases, these data also establish a novel class of nucleases.
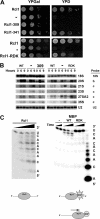
Figures



Similar articles
-
Maize RNA 3'-terminal phosphate cyclase-like protein promotes 18S pre-rRNA cleavage and is important for kernel development.Plant Cell. 2022 Apr 26;34(5):1957-1979. doi: 10.1093/plcell/koac052. Plant Cell. 2022. PMID: 35167702 Free PMC article.
-
Rcl1p, the yeast protein similar to the RNA 3'-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis.EMBO J. 2000 May 2;19(9):2115-26. doi: 10.1093/emboj/19.9.2115. EMBO J. 2000. PMID: 10790377 Free PMC article.
-
An RNA conformational switch regulates pre-18S rRNA cleavage.J Mol Biol. 2011 Jan 7;405(1):3-17. doi: 10.1016/j.jmb.2010.09.064. Epub 2010 Oct 8. J Mol Biol. 2011. PMID: 20934433 Free PMC article.
-
Caught in the act-Visualizing ribonucleases during eukaryotic ribosome assembly.Wiley Interdiscip Rev RNA. 2023 Jul-Aug;14(4):e1766. doi: 10.1002/wrna.1766. Epub 2022 Oct 18. Wiley Interdiscip Rev RNA. 2023. PMID: 36254602 Review.
-
Processing of pre-ribosomal RNA in Saccharomyces cerevisiae.Yeast. 1995 Dec;11(16):1629-50. doi: 10.1002/yea.320111607. Yeast. 1995. PMID: 8720068 Review.
Cited by
-
Maize RNA 3'-terminal phosphate cyclase-like protein promotes 18S pre-rRNA cleavage and is important for kernel development.Plant Cell. 2022 Apr 26;34(5):1957-1979. doi: 10.1093/plcell/koac052. Plant Cell. 2022. PMID: 35167702 Free PMC article.
-
Identifying Prognostic Significance of RCL1 and Four-Gene Signature as Novel Potential Biomarkers in HCC Patients.J Oncol. 2021 Jun 28;2021:5574150. doi: 10.1155/2021/5574150. eCollection 2021. J Oncol. 2021. PMID: 34257652 Free PMC article.
-
Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18S rRNA.Nucleic Acids Res. 2013 Apr;41(8):4709-23. doi: 10.1093/nar/gkt160. Epub 2013 Mar 12. Nucleic Acids Res. 2013. PMID: 23482395 Free PMC article.
-
Rrp5 establishes a checkpoint for 60S assembly during 40S maturation.RNA. 2019 Sep;25(9):1164-1176. doi: 10.1261/rna.071225.119. Epub 2019 Jun 19. RNA. 2019. PMID: 31217256 Free PMC article.
-
The small and large ribosomal subunits depend on each other for stability and accumulation.Life Sci Alliance. 2019 Mar 5;2(2):e201800150. doi: 10.26508/lsa.201800150. Print 2019 Apr. Life Sci Alliance. 2019. PMID: 30837296 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

