Discovery and preclinical validation of drug indications using compendia of public gene expression data
- PMID: 21849665
- PMCID: PMC3502016
- DOI: 10.1126/scitranslmed.3001318
Discovery and preclinical validation of drug indications using compendia of public gene expression data
Erratum in
- Sci Transl Med. 2011 Sep 28;3(102):102er7
Abstract
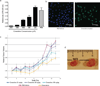
The application of established drug compounds to new therapeutic indications, known as drug repositioning, offers several advantages over traditional drug development, including reduced development costs and shorter paths to approval. Recent approaches to drug repositioning use high-throughput experimental approaches to assess a compound's potential therapeutic qualities. Here, we present a systematic computational approach to predict novel therapeutic indications on the basis of comprehensive testing of molecular signatures in drug-disease pairs. We integrated gene expression measurements from 100 diseases and gene expression measurements on 164 drug compounds, yielding predicted therapeutic potentials for these drugs. We recovered many known drug and disease relationships using computationally derived therapeutic potentials and also predict many new indications for these 164 drugs. We experimentally validated a prediction for the antiulcer drug cimetidine as a candidate therapeutic in the treatment of lung adenocarcinoma, and demonstrate its efficacy both in vitro and in vivo using mouse xenograft models. This computational method provides a systematic approach for repositioning established drugs to treat a wide range of human diseases.
Figures




Comment in
-
The emergence of genome-based drug repositioning.Sci Transl Med. 2011 Aug 17;3(96):96ps35. doi: 10.1126/scitranslmed.3001512. Sci Transl Med. 2011. PMID: 21849663 Free PMC article.
-
Genetic signatures uncover new uses.Nat Rev Drug Discov. 2011 Sep 16;10(10):732-3. doi: 10.1038/nrd3565. Nat Rev Drug Discov. 2011. PMID: 21921920 No abstract available.
-
Signatures for drug repositioning.Nat Rev Genet. 2011 Sep 16;12(10):668. doi: 10.1038/nrg3076. Nat Rev Genet. 2011. PMID: 21921924 No abstract available.
References
-
- Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004 Aug;3:673. - PubMed
-
- DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. J Health Econ. 2003 Mar;22:151. - PubMed
-
- Chong CR, Sullivan DJ., Jr New uses for old drugs. Nature. 2007 Aug 9;448:645. - PubMed
-
- Lum PY, Derry JM, Schadt EE. Integrative genomics and drug development. Pharmacogenomics. 2009 Feb;10:203. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

