Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes
- PMID: 21849976
- PMCID: PMC3257054
- DOI: 10.1038/nature10378
Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes
Abstract
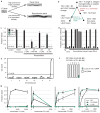
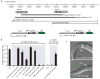
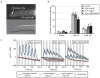
Evolution can follow predictable genetic trajectories, indicating that discrete environmental shifts can select for reproducible genetic changes. Conspecific individuals are an important feature of an animal's environment, and a potential source of selective pressures. Here we show that adaptation of two Caenorhabditis species to growth at high density, a feature common to domestic environments, occurs by reproducible genetic changes to pheromone receptor genes. Chemical communication through pheromones that accumulate during high-density growth causes young nematode larvae to enter the long-lived but non-reproductive dauer stage. Two strains of Caenorhabditis elegans grown at high density have independently acquired multigenic resistance to pheromone-induced dauer formation. In each strain, resistance to the pheromone ascaroside C3 results from a deletion that disrupts the adjacent chemoreceptor genes serpentine receptor class g (srg)-36 and -37. Through misexpression experiments, we show that these genes encode redundant G-protein-coupled receptors for ascaroside C3. Multigenic resistance to dauer formation has also arisen in high-density cultures of a different nematode species, Caenorhabditis briggsae, resulting in part from deletion of an srg gene paralogous to srg-36 and srg-37. These results demonstrate rapid remodelling of the chemoreceptor repertoire as an adaptation to specific environments, and indicate that parallel changes to a common genetic substrate can affect life-history traits across species.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

