Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers
- PMID: 21850056
- PMCID: PMC3260502
- DOI: 10.1038/ismej.2011.109
Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers
Abstract
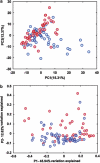
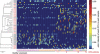
Despite a long-suspected role in the development of human colorectal cancer (CRC), the composition of gut microbiota in CRC patients has not been adequately described. In this study, fecal bacterial diversity in CRC patients (n=46) and healthy volunteers (n=56) were profiled by 454 pyrosequencing of the V3 region of the 16S ribosomal RNA gene. Both principal component analysis and UniFrac analysis showed structural segregation between the two populations. Forty-eight operational taxonomic units (OTUs) were identified by redundancy analysis as key variables significantly associated with the structural difference. One OTU closely related to Bacteroides fragilis was enriched in the gut microbiota of CRC patients, whereas three OTUs related to Bacteroides vulgatus and Bacteroides uniformis were enriched in that of healthy volunteers. A total of 11 OTUs belonging to the genera Enterococcus, Escherichia/Shigella, Klebsiella, Streptococcus and Peptostreptococcus were significantly more abundant in the gut microbiota of CRC patients, and 5 OTUs belonging to the genus Roseburia and other butyrate-producing bacteria of the family Lachnospiraceae were less abundant. Real-time quantitative PCR further validated the significant reduction of butyrate-producing bacteria in the gut microbiota of CRC patients by measuring the copy numbers of butyryl-coenzyme A CoA transferase genes (Mann-Whitney test, P<0.01). Reduction of butyrate producers and increase of opportunistic pathogens may constitute a major structural imbalance of gut microbiota in CRC patients.
Figures

References
-
- Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23 (Part 1:1298–1303. - PubMed
-
- Biarc J, Nguyen IS, Pini A, Gosse F, Richert S, Thierse D, et al. Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S.bovis) Carcinogenesis. 2004;25:1477–1484. - PubMed
-
- Bingham SA. Diet and colorectal cancer prevention. Biochem Soc Trans. 2000;28:12–16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

