RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function
- PMID: 21852534
- PMCID: PMC3165934
- DOI: 10.1101/gad.16651211
RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function
Abstract
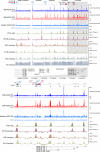
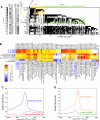
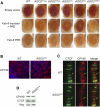
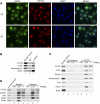
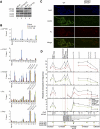
A major role of the RNAi pathway in Schizosaccharomyces pombe is to nucleate heterochromatin, but it remains unclear whether this mechanism is conserved. To address this question in Drosophila, we performed genome-wide localization of Argonaute2 (AGO2) by chromatin immunoprecipitation (ChIP)-seq in two different embryonic cell lines and found that AGO2 localizes to euchromatin but not heterochromatin. This localization pattern is further supported by immunofluorescence staining of polytene chromosomes and cell lines, and these studies also indicate that a substantial fraction of AGO2 resides in the nucleus. Intriguingly, AGO2 colocalizes extensively with CTCF/CP190 chromatin insulators but not with genomic regions corresponding to endogenous siRNA production. Moreover, AGO2, but not its catalytic activity or Dicer-2, is required for CTCF/CP190-dependent Fab-8 insulator function. AGO2 interacts physically with CTCF and CP190, and depletion of either CTCF or CP190 results in genome-wide loss of AGO2 chromatin association. Finally, mutation of CTCF, CP190, or AGO2 leads to reduction of chromosomal looping interactions, thereby altering gene expression. We propose that RNAi-independent recruitment of AGO2 to chromatin by insulator proteins promotes the definition of transcriptional domains throughout the genome.
Figures
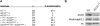

References
-
- Amero SA, Elgin SC, Beyer AL 1991. A unique zinc finger protein is associated preferentially with active ecdysone-responsive loci in Drosophila. Genes Dev 5: 188–200 - PubMed
-
- Bailey TL, Elkan C 1995. The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3: 21–29 - PubMed
-
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G 2011. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144: 214–226 - PubMed
-
- Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F 2000. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127: 779–790 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
