Paraoxonase (PON1 and PON3) Polymorphisms: Impact on Liver Expression and Atorvastatin-Lactone Hydrolysis
- PMID: 21852972
- PMCID: PMC3147178
- DOI: 10.3389/fphar.2011.00041
Paraoxonase (PON1 and PON3) Polymorphisms: Impact on Liver Expression and Atorvastatin-Lactone Hydrolysis
Abstract
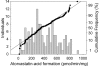
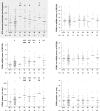
Atorvastatin δ-lactone, a major, pharmacologically inactive metabolite, has been associated with toxicity. In a previous study we showed that polymorphisms of UGT1A3 influence atorvastatin δ-lactone formation. Here we investigated the reverse reaction, atorvastatin δ-lactone hydrolysis, in a human liver bank. Screening of microarray data revealed paraoxonases PON1 and PON3 among 17 candidate esterases. Microsomal δ-lactone hydrolysis was significantly correlated to PON1 and PON3 protein (r(s) = 0.60; r(s) = 0.62, respectively; P < 0.0001). PON1 and PON3 were strongly correlated to each other (r(s) = 0.60) but PON1 was shown to be more extensively glycosylated than PON3. In addition a novel splice-variant of PON3 was identified. Genotyping of 40 polymorphisms within the PON-locus identified PON1 promoter polymorphisms (-108T > C, -832G > A, -1741G > A) and a tightly linked group of PON3 polymorphisms (-4984A > G, -4105G > A, -1091A > G, -746C > T, and F21F) to be associated with changes in atorvastatin δ-lactone hydrolysis and expression of PON1 but not PON3. However, carriers of the common PON1 polymorphisms L55M or Q192R showed no difference in δ-lactone hydrolysis or PON expression. Haplotype analysis revealed decreased δ-lactone hydrolysis in carriers of the most common haplotype *1 compared to carriers of haplotypes *2, *3, *4, and *7. Analysis of non-genetic factors showed association of hepatocellular and cholangiocellular carcinoma with decreased PON1 and PON3 expression, respectively. Increased C-reactive protein and γ-glutamyl transferase levels were associated with decreased protein expression of both enzymes, and increased bilirubin levels, cholestasis, and presurgical exposure to omeprazole or pantoprazole were related to decreased PON3 protein. In conclusion, PON-locus polymorphisms affect PON1 expression whereas non-genetic factors have an effect on PON1 and PON3 expression. This may influence response to therapy or adverse events in statin treatment.
Keywords: PON1; PON3; SNP; atorvastatin-lactone; myopathy; paraoxonase; pharmacogenetics; rhabdomyolysis.
Figures
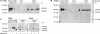




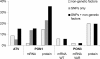

References
-
- Aragones G., Guardiola M., Barreda M., Marsillach J., Beltran-Debon R., Rull A., Mackness B., Mackness M., Joven J., Simo J. M., Camps J. (2011). Measurement of serum paraoxonase-3 concentration: method evaluation, reference values and influence of genotypes in a population-based study. J. Lipid Res. 52, 1055–1061 10.1194/jlr.D014134 - DOI - PMC - PubMed
-
- Baer A. N., Wortmann R. L. (2007). Myotoxicity associated with lipid-lowering drugs. Curr. Opin. Rheumatol. 19, 67–73 - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

