Characterization of the MDSC proteome associated with metastatic murine mammary tumors using label-free mass spectrometry and shotgun proteomics
- PMID: 21853032
- PMCID: PMC3154190
- DOI: 10.1371/journal.pone.0022446
Characterization of the MDSC proteome associated with metastatic murine mammary tumors using label-free mass spectrometry and shotgun proteomics
Abstract
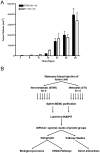
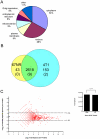
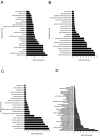
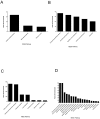
Expansion of Gr-1+/CD11b+ myeloid derived suppressor cells (MDSCs) is governed by the presence of increasingly metastatic, malignant primary tumors. Metastasis, not the primary tumor, is often the cause of mortality. This study sought to fully characterize the MDSC proteome in response to metastatic and non-metastatic mammary tumors using label-free mass spectrometry shotgun proteomics in a mouse model with tumor cell lines, 67NR and 4T1, derived from the same tumor. 67NR cells form only primary mammary tumors, whereas 4T1 cells readily metastasize to the lungs, lymph nodes, and blood. Overall analysis identified a total of 2825 protein groups with a 0.78% false discovery rate. Of the 2814 true identifications, 43 proteins were exclusive to the 67NR group, 153 were exclusive to the 4T1 group, and 2618 were shared. Among the shared cohort, 26 proteins were increased and 31 were decreased in the metastatic 4T1 cohort compared to non-metastatic 67NR controls after filtering. MDSCs selectively express proteins involved in the γ-glutamyl transferase, glutathione synthase pathways, CREB transcription factor signaling, and other pathways involved in platelet aggregation, as well as lipid and amino acid metabolism, in response to highly metastatic 4T1 tumors. Cell cycle regulation dominated protein pathways and ontological groups of the 67NR non-metastatic group. Not only does this study provide a starting point to identify potential biomarkers of metastasis expressed by MDSCs; it identifies critical pathways that are unique to non-metastatic and metastatic conditions. Therapeutic interventions aimed at these pathways in MDSC may offer a new route to control malignancy and metastasis.
Conflict of interest statement
Figures
References
-
- Ribechini E, Greifenberg V, Sandwick S, Lutz MB. Subsets, expansion and activation of myeloid-derived suppressor cells. Med Microbiol Immunol. 2010;199:273–281. - PubMed
-
- Almand B, Clark JI, Nikitina E, van Beynen J, English NR, et al. Increased production of immature myeloid cells in cancer patients: a mechanism of immunosuppression in cancer. J Immunol. 2001;166:678–689. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

