Comparative analysis of salivary bacterial microbiome diversity in edentulous infants and their mothers or primary care givers using pyrosequencing
- PMID: 21853142
- PMCID: PMC3154475
- DOI: 10.1371/journal.pone.0023503
Comparative analysis of salivary bacterial microbiome diversity in edentulous infants and their mothers or primary care givers using pyrosequencing
Abstract
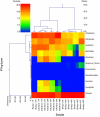
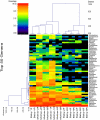
Bacterial contribution to oral disease has been studied in young children, but there is a lack of data addressing the developmental perspective in edentulous infants. Our primary objectives were to use pyrosequencing to phylogenetically characterize the salivary bacterial microbiome of edentulous infants and to make comparisons against their mothers. Saliva samples were collected from 5 edentulous infants (mean age = 4.6±1.2 mo old) and their mothers or primary care givers (mean age = 30.8±9.5 y old). Salivary DNA was extracted, used to generate DNA amplicons of the V4-V6 hypervariable region of the bacterial 16S rDNA gene, and subjected to 454-pyrosequencing. On average, over 80,000 sequences per sample were generated. High bacterial diversity was noted in the saliva of adults [1012 operational taxonomical units (OTU) at 3% divergence] and infants (578 OTU at 3% divergence). Firmicutes, Proteobacteria, Actinobacteria, and Fusobacteria were predominant bacterial phyla present in all samples. A total of 397 bacterial genera were present in our dataset. Of the 28 genera different (P<0.05) between infants and adults, 27 had a greater prevalence in adults. The exception was Streptococcus, which was the predominant genera in infant saliva (62.2% in infants vs. 20.4% in adults; P<0.05). Veillonella, Neisseria, Rothia, Haemophilus, Gemella, Granulicatella, Leptotrichia, and Fusobacterium were also predominant genera in infant samples, while Haemophilus, Neisseria, Veillonella, Fusobacterium, Oribacterium, Rothia, Treponema, and Actinomyces were predominant in adults. Our data demonstrate that although the adult saliva bacterial microbiome had a greater OTU count than infants, a rich bacterial community exists in the infant oral cavity prior to tooth eruption. Streptococcus, Veillonella, and Neisseria are the predominant bacterial genera present in infants. Further research is required to characterize the development of oral microbiota early in life and identify environmental factors that impact colonization and oral and gastrointestinal disease risk.
Conflict of interest statement
Figures
References
-
- Beltran-Aguilar ED, Barker LK, Canto MT, Dye BA, Gooch BF, et al. Surveillance for dental caries, dental sealants, tooth retention, edentulism, and enamel fluorosis – United States, 1988-1994 and 1999-2002. MMWR Surveill Summ. 2005;54:1–43. - PubMed
-
- Socransky SS, Haffajee DD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. - PubMed
-
- Sanchez-Perez L, Golubov J, Irigoyen-Camacho ME, Moctezuma PA, Acosta-Gio E. Clinical, salivary, and bacterial markers for caries risk assessment in schoolchildren: a 4-year follow-up. Int J Paed Dent. 2009;19:186–192. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

