Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development
- PMID: 21854559
- PMCID: PMC3277949
- DOI: 10.1186/gb-2011-12-8-r81
Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development
Erratum in
- Genome Biol. 2011;12(12):414. Patel, Hardip R [added]
Abstract
Background: We present the genome sequence of the tammar wallaby, Macropus eugenii, which is a member of the kangaroo family and the first representative of the iconic hopping mammals that symbolize Australia to be sequenced. The tammar has many unusual biological characteristics, including the longest period of embryonic diapause of any mammal, extremely synchronized seasonal breeding and prolonged and sophisticated lactation within a well-defined pouch. Like other marsupials, it gives birth to highly altricial young, and has a small number of very large chromosomes, making it a valuable model for genomics, reproduction and development.
Results: The genome has been sequenced to 2 × coverage using Sanger sequencing, enhanced with additional next generation sequencing and the integration of extensive physical and linkage maps to build the genome assembly. We also sequenced the tammar transcriptome across many tissues and developmental time points. Our analyses of these data shed light on mammalian reproduction, development and genome evolution: there is innovation in reproductive and lactational genes, rapid evolution of germ cell genes, and incomplete, locus-specific X inactivation. We also observe novel retrotransposons and a highly rearranged major histocompatibility complex, with many class I genes located outside the complex. Novel microRNAs in the tammar HOX clusters uncover new potential mammalian HOX regulatory elements.
Conclusions: Analyses of these resources enhance our understanding of marsupial gene evolution, identify marsupial-specific conserved non-coding elements and critical genes across a range of biological systems, including reproduction, development and immunity, and provide new insight into marsupial and mammalian biology and genome evolution.
Figures
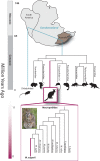
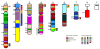
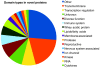
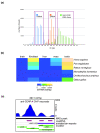
Comment in
-
Sequencing skippy: the genome sequence of an Australian kangaroo, Macropus eugenii.Genome Biol. 2011 Aug 19;12(8):123. doi: 10.1186/gb-2011-12-8-123. Genome Biol. 2011. PMID: 21861852 Free PMC article.
References
-
- Drake-Brockman H. Voyage to Disaster. Sydney: Angus and Robertson; 1963.
-
- Meredith RW, Westerman M, Springer MS. A phylogeny and timescale for the living genera of kangaroos and kin (Macropodiformes: Marsupialia) based on nuclear DNA sequences. Aust J Zool. 2008;56:395–410. doi: 10.1071/ZO08044. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

