Characterisation of marsupial PHLDA2 reveals eutherian specific acquisition of imprinting
- PMID: 21854573
- PMCID: PMC3170258
- DOI: 10.1186/1471-2148-11-244
Characterisation of marsupial PHLDA2 reveals eutherian specific acquisition of imprinting
Abstract
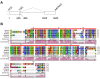
Background: Genomic imprinting causes parent-of-origin specific gene expression by differential epigenetic modifications between two parental genomes. We previously reported that there is no evidence of genomic imprinting of CDKN1C in the KCNQ1 domain in the placenta of an Australian marsupial, the tammar wallaby (Macropus eugenii) whereas tammar IGF2 and H19, located adjacent to the KCNQ1 domain in eutherian mammals, are imprinted. We have now identified and characterised the marsupial orthologue of PHLDA2, another gene in the KCNQ1 domain (also known as IPL or TSSC3) that is imprinted in eutherians. In mice, Phlda2 is a dose-sensitive negative regulator of placental growth, as Cdkn1c is for embryonic growth.
Results: Tammar PHLDA2 is highly expressed in the yolk sac placenta compared to other fetal tissues, confirming a similar expression pattern to that of mouse Phlda2. However, tammar PHLDA2 is biallelically expressed in both the fetus and yolk sac placenta, so it is not imprinted. The lack of imprinting in tammar PHLDA2 suggests that the acquisition of genomic imprinting of the KCNQ1 domain in eutherian mammals, accompanied with gene dosage reduction, occurred after the split of the therian mammals into the marsupials and eutherians.
Conclusions: Our results confirm the idea that acquisition of genomic imprinting in the KCNQ1 domain occurred specifically in the eutherian lineage after the divergence of marsupials, even though imprinting of the adjacent IGF2-H19 domain arose before the marsupial-eutherian split. These data are consistent with the hypothesis that genomic imprinting of the KCNQ1 domain may have contributed to the evolution of more complex placentation in the eutherian lineage by reduction of the gene dosage of negative regulators for both embryonic and placental growth.
Figures
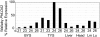


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

