Degeneration of penicillin production in ethanol-limited chemostat cultivations of Penicillium chrysogenum: A systems biology approach
- PMID: 21854586
- PMCID: PMC3224390
- DOI: 10.1186/1752-0509-5-132
Degeneration of penicillin production in ethanol-limited chemostat cultivations of Penicillium chrysogenum: A systems biology approach
Abstract
Background: In microbial production of non-catabolic products such as antibiotics a loss of production capacity upon long-term cultivation (for example chemostat), a phenomenon called strain degeneration, is often observed. In this study a systems biology approach, monitoring changes from gene to produced flux, was used to study degeneration of penicillin production in a high producing Penicillium chrysogenum strain during prolonged ethanol-limited chemostat cultivations.
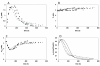
Results: During these cultivations, the biomass specific penicillin production rate decreased more than 10-fold in less than 22 generations. No evidence was obtained for a decrease of the copy number of the penicillin gene cluster, nor a significant down regulation of the expression of the penicillin biosynthesis genes. However, a strong down regulation of the biosynthesis pathway of cysteine, one of the precursors of penicillin, was observed. Furthermore the protein levels of the penicillin pathway enzymes L-α-(δ-aminoadipyl)-L-α-cystenyl-D-α-valine synthetase (ACVS) and isopenicillin-N synthase (IPNS), decreased significantly. Re-cultivation of fully degenerated cells in unlimited batch culture and subsequent C-limited chemostats did only result in a slight recovery of penicillin production.
Conclusions: Our findings indicate that the observed degeneration is attributed to a significant decrease of the levels of the first two enzymes of the penicillin biosynthesis pathway, ACVS and IPNS. This decrease is not caused by genetic instability of the penicillin amplicon, neither by down regulation of the penicillin biosynthesis pathway. Furthermore no indications were obtained for degradation of these enzymes as a result of autophagy. Possible causes for the decreased enzyme levels could be a decrease of the translation efficiency of ACVS and IPNS during degeneration, or the presence of a culture variant impaired in the biosynthesis of functional proteins of these enzymes, which outcompeted the high producing part of the population.
Figures

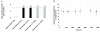
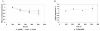



References
-
- Fleming A. Antibacterial Action of Cultures of A Penicillium, with Special Reference to Their Use in the Isolation of B. Influenza. Br J Exp Pathol. 1928;10:185–194.
-
- Newbert RW, Barton B, Greaves P, Harper J, Turner G. Analysis of a commercially improved Penicillium chrysogenum strain series: involvement of recombinogenic regions in amplification and deletion of the penicillin biosynthesis gene cluster. J Ind Microbiol Biotechnol. 1997;19:18–27. doi: 10.1038/sj.jim.2900411. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

