The tammar wallaby major histocompatibility complex shows evidence of past genomic instability
- PMID: 21854592
- PMCID: PMC3179965
- DOI: 10.1186/1471-2164-12-421
The tammar wallaby major histocompatibility complex shows evidence of past genomic instability
Abstract
Background: The major histocompatibility complex (MHC) is a group of genes with a variety of roles in the innate and adaptive immune responses. MHC genes form a genetically linked cluster in eutherian mammals, an organization that is thought to confer functional and evolutionary advantages to the immune system. The tammar wallaby (Macropus eugenii), an Australian marsupial, provides a unique model for understanding MHC gene evolution, as many of its antigen presenting genes are not linked to the MHC, but are scattered around the genome.
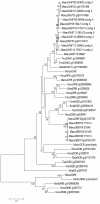
Results: Here we describe the 'core' tammar wallaby MHC region on chromosome 2q by ordering and sequencing 33 BAC clones, covering over 4.5 MB and containing 129 genes. When compared to the MHC region of the South American opossum, eutherian mammals and non-mammals, the wallaby MHC has a novel gene organization. The wallaby has undergone an expansion of MHC class II genes, which are separated into two clusters by the class III genes. The antigen processing genes have undergone duplication, resulting in two copies of TAP1 and three copies of TAP2. Notably, Kangaroo Endogenous Retroviral Elements are present within the region and may have contributed to the genomic instability.
Conclusions: The wallaby MHC has been extensively remodeled since the American and Australian marsupials last shared a common ancestor. The instability is characterized by the movement of antigen presenting genes away from the core MHC, most likely via the presence and activity of retroviral elements. We propose that the movement of class II genes away from the ancestral class II region has allowed this gene family to expand and diversify in the wallaby. The duplication of TAP genes in the wallaby MHC makes this species a unique model organism for studying the relationship between MHC gene organization and function.
Figures





References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

