Single-cell copy number variation detection
- PMID: 21854607
- PMCID: PMC3245619
- DOI: 10.1186/gb-2011-12-8-r80
Single-cell copy number variation detection
Abstract
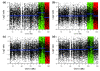
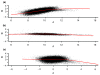
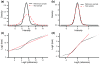
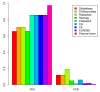
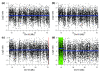
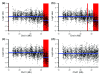
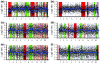
Detection of chromosomal aberrations from a single cell by array comparative genomic hybridization (single-cell array CGH), instead of from a population of cells, is an emerging technique. However, such detection is challenging because of the genome artifacts and the DNA amplification process inherent to the single cell approach. Current normalization algorithms result in inaccurate aberration detection for single-cell data. We propose a normalization method based on channel, genome composition and recurrent genome artifact corrections. We demonstrate that the proposed channel clone normalization significantly improves the copy number variation detection in both simulated and real single-cell array CGH data.
Figures

References
-
- Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit F, Fryns J-P, Verbeke G, D'Hooghe T, Moreau Y, Vermeesch JR. Chromosome instability is common in human cleavage-stage embryos. Nature medicine. 2009;15:577–583. doi: 10.1038/nm.1924. - DOI - PubMed
-
- Handyside AH, Harton GL, Mariani B, Thornhill AR, Affara N, Shaw M-A, Griffin DK. Karyomapping: a universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. Journal of medical genetics. 2010;47:651–658. doi: 10.1136/jmg.2009.069971. - DOI - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

