Specific roles for the GATA transcription factors end-1 and end-3 during C. elegans E-lineage development
- PMID: 21854766
- PMCID: PMC4454387
- DOI: 10.1016/j.ydbio.2011.08.002
Specific roles for the GATA transcription factors end-1 and end-3 during C. elegans E-lineage development
Abstract
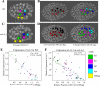
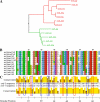
end-1 and end-3 are GATA transcription factors important for specifying endoderm cell fate in Caenorhabditis elegans. Deletion of both factors together results in larval arrest, 0% survival and a fate change in the endoderm-specifying E lineage. Individual deletions of either factor, however, result in the development of viable, fertile adults, with 100% of worms developing to adults for end-1(-) and 95% for end-3(-). We sought to quantify the variable phenotypes seen in both deletions using automated cell lineaging. We quantified defects in cell lifetime, cell movement and division axis in end-3(-) embryos, while quantifying perturbations in downstream reporter gene expression in strains with homozygous deletions for either gene, showing that each deletion leads to a unique profile of downstream perturbations in gene expression and cellular phenotypes with a high correlation between early and late defects. Combining observations in both cellular and gene expression defects we found that misaligned divisions at the E2 stage resulted in ectopic expression of the Notch target ref-1 in end-3(-) embryos. Using a maximum likelihood phylogenetic approach we found end-1 and end-3 split to form two distinct clades within the Caenorhabditis lineage with distinct DNA-binding structures. These results indicate that end-1 and end-3 have each evolved into genes with unique functions during endoderm development, that end-3(-) embryos have a delay in the onset of E lineage cell fate and that end-1 has only a partially penetrant ability to activate E lineage fate.
Copyright © 2011. Published by Elsevier Inc.
Figures

References
-
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–552. - PubMed
-
- Baugh LR, Hill AA, Slonim AK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130(5):889–900. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

