Mechanisms for the inheritance of chromatin states
- PMID: 21854979
- PMCID: PMC3244757
- DOI: 10.1016/j.cell.2011.07.013
Mechanisms for the inheritance of chromatin states
Abstract
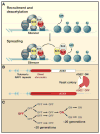
Studies in eukaryotes ranging from yeast to mammals indicate that specific chromatin structures can be inherited following DNA replication via mechanisms acting in cis. Both the initial establishment of such chromatin structures and their inheritance require sequence-dependent specificity factors and changes in histone posttranslational modifications. Here I propose models for the maintenance of epigenetic information in which DNA silencers or nascent RNA scaffolds act as sensors that work cooperatively with parentally inherited histones to re-establish chromatin states following DNA replication.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



References
-
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. - PubMed
-
- Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet. 2011;12:123–135. - PubMed
-
- Bell SP, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

