The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly
- PMID: 21854981
- PMCID: PMC3291198
- DOI: 10.1016/j.cell.2011.07.012
The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly
Abstract
Error-free chromosome segregation requires stable attachment of sister kinetochores to the opposite spindle poles (amphitelic attachment). Exactly how amphitelic attachments are achieved during spindle assembly remains elusive. We employed photoactivatable GFP and high-resolution live-cell confocal microscopy to visualize complete 3D movements of individual kinetochores throughout mitosis in nontransformed human cells. Combined with electron microscopy, molecular perturbations, and immunofluorescence analyses, this approach reveals unexpected details of chromosome behavior. Our data demonstrate that unstable lateral interactions between kinetochores and microtubules dominate during early prometaphase. These transient interactions lead to the reproducible arrangement of chromosomes in an equatorial ring on the surface of the nascent spindle. A computational model predicts that this toroidal distribution of chromosomes exposes kinetochores to a high density of microtubules which facilitates subsequent formation of amphitelic attachments. Thus, spindle formation involves a previously overlooked stage of chromosome prepositioning which promotes formation of amphitelic attachments.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

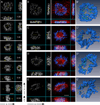



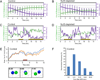

Comment in
-
Travel notes from the equatorial circle.Cell. 2011 Aug 19;146(4):499-501. doi: 10.1016/j.cell.2011.08.001. Cell. 2011. PMID: 21854975
-
Cell cycle: the division belt.Nat Rev Mol Cell Biol. 2011 Sep 14;12(10):622. doi: 10.1038/nrm3194. Nat Rev Mol Cell Biol. 2011. PMID: 21915142 No abstract available.
References
-
- Blangy A, Lane HA, d'Herin P, Harper M, Kress M, Nigg EA. Phosphorilation by p34 cdc2 regulates spindle assosiation of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

