Identification of a rapidly formed nonnucleosomal histone-DNA intermediate that is converted into chromatin by ACF
- PMID: 21855802
- PMCID: PMC3160715
- DOI: 10.1016/j.molcel.2011.07.017
Identification of a rapidly formed nonnucleosomal histone-DNA intermediate that is converted into chromatin by ACF
Abstract
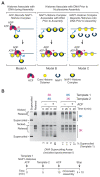
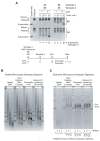
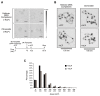
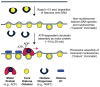
Chromatin assembly involves the combined action of histone chaperones and ATP-dependent motor proteins. Here, we investigate the mechanism of nucleosome assembly with a purified chromatin assembly system containing the histone chaperone NAP1 and the ATP-dependent motor protein ACF. These studies revealed the rapid formation of a stable nonnucleosomal histone-DNA intermediate that is converted into canonical nucleosomes by ACF. The histone-DNA intermediate does not supercoil DNA like a canonical nucleosome, but has a nucleosome-like appearance by atomic force microscopy. This intermediate contains all four core histones, lacks NAP1, and is formed by the initial deposition of histones H3-H4. Conversion of the intermediate into histone H1-containing chromatin results in increased resistance to micrococcal nuclease digestion. These findings suggest that the histone-DNA intermediate corresponds to nascent nucleosome-like structures, such as those observed at DNA replication forks. Related complexes might be formed during other chromatin-directed processes such as transcription, DNA repair, and histone exchange.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
The prenucleosome, a stable conformational isomer of the nucleosome.Genes Dev. 2015 Dec 15;29(24):2563-75. doi: 10.1101/gad.272633.115. Genes Dev. 2015. PMID: 26680301 Free PMC article.
-
The histone chaperones Nap1 and Vps75 bind histones H3 and H4 in a tetrameric conformation.Mol Cell. 2011 Feb 18;41(4):398-408. doi: 10.1016/j.molcel.2011.01.025. Mol Cell. 2011. PMID: 21329878 Free PMC article.
-
Structural plasticity of histones H3-H4 facilitates their allosteric exchange between RbAp48 and ASF1.Nat Struct Mol Biol. 2013 Jan;20(1):29-35. doi: 10.1038/nsmb.2446. Epub 2012 Nov 25. Nat Struct Mol Biol. 2013. PMID: 23178455 Free PMC article.
-
All roads lead to chromatin: multiple pathways for histone deposition.Biochim Biophys Acta. 2013 Mar-Apr;1819(3-4):238-46. Biochim Biophys Acta. 2013. PMID: 24459726 Review.
-
Prenucleosomes and Active Chromatin.Cold Spring Harb Symp Quant Biol. 2015;80:65-72. doi: 10.1101/sqb.2015.80.027300. Epub 2016 Jan 14. Cold Spring Harb Symp Quant Biol. 2015. PMID: 26767995 Free PMC article. Review.
Cited by
-
Chromatin and DNA replication.Cold Spring Harb Perspect Biol. 2013 Aug 1;5(8):a010207. doi: 10.1101/cshperspect.a010207. Cold Spring Harb Perspect Biol. 2013. PMID: 23751185 Free PMC article. Review.
-
Molecular basis for chromatin assembly and modification by multiprotein complexes.Protein Sci. 2019 Feb;28(2):329-343. doi: 10.1002/pro.3535. Epub 2018 Dec 13. Protein Sci. 2019. PMID: 30350439 Free PMC article. Review.
-
ATRX limits the accessibility of histone H3-occupied HSV genomes during lytic infection.PLoS Pathog. 2021 Apr 28;17(4):e1009567. doi: 10.1371/journal.ppat.1009567. eCollection 2021 Apr. PLoS Pathog. 2021. PMID: 33909709 Free PMC article.
-
Active promoters give rise to false positive 'Phantom Peaks' in ChIP-seq experiments.Nucleic Acids Res. 2015 Aug 18;43(14):6959-68. doi: 10.1093/nar/gkv637. Epub 2015 Jun 27. Nucleic Acids Res. 2015. PMID: 26117547 Free PMC article.
-
Replication-Coupled Nucleosome Assembly and Positioning by ATP-Dependent Chromatin-Remodeling Enzymes.Cell Rep. 2016 Apr 26;15(4):715-723. doi: 10.1016/j.celrep.2016.03.059. Epub 2016 Apr 14. Cell Rep. 2016. PMID: 27149855 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

