Characterization of DOK1, a candidate tumor suppressor gene, in epithelial ovarian cancer
- PMID: 21856257
- PMCID: PMC5528302
- DOI: 10.1016/j.molonc.2011.07.003
Characterization of DOK1, a candidate tumor suppressor gene, in epithelial ovarian cancer
Abstract
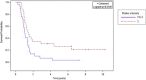
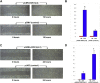
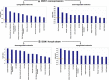
In attempt to discover novel aberrantly hypermethylated genes with putative tumor suppressor function in epithelial ovarian cancer (EOC), we applied expression profiling following pharmacologic inhibition of DNA methylation in EOC cell lines. Among the genes identified, one of particular interest was DOK1, or downstream of tyrosine kinase 1, previously recognized as a candidate tumor suppressor gene (TSG) for leukemia and other human malignancies. Using bisulfite sequencing, we determined that a 5'-non-coding DNA region (located at nt -1158 to -850, upstream of the DOK1 translation start codon) was extensively hypermethylated in primary serous EOC tumors compared with normal ovarian specimens; however, this hypermethylation was not associated with DOK1 suppression. On the contrary, DOK1 was found to be strongly overexpressed in serous EOC tumors as compared to normal tissue and importantly, DOK1 overexpression significantly correlated with improved progression-free survival (PFS) values of serous EOC patients. Ectopic modulation of DOK1 expression in EOC cells and consecutive functional analyses pointed toward association of DOK1 expression with increased EOC cell migration and proliferation, and better sensitivity to cisplatin treatment. Gene expression profiling and consecutive network and pathway analyses were also confirmative for DOK1 association with EOC cell migration and proliferation. These analyses were also indicative for DOK1 protective role in EOC tumorigenesis, linked to DOK1-mediated induction of some tumor suppressor factors and its suppression of pro-metastasis genes. Taken together, our findings are suggestive for a possible tumor suppressor role of DOK1 in EOC; however its implication in enhanced EOC cell migration and proliferation restrain us to conclude that DOK1 represents a true TSG in EOC. Further studies are needed to more completely elucidate the functional implications of DOK1 and other members of the DOK gene family in ovarian tumorigenesis.
Copyright © 2011 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures






References
-
- Bachvarov, D. , L'Esperance, S. , Popa, I. , Bachvarova, M. , Plante, M. , Tetu, B. , 2006. Gene expression patterns of chemoresistant and chemosensitive serous epithelial ovarian tumors with possible predictive value in response to initial chemotherapy. International Journal of Oncology. 29, 919–933. - PubMed
-
- Balch, C. , Huang, T.H. , Brown, R. , Nephew, K.P. , 2004. The epigenetics of ovarian cancer drug resistance and resensitization. American Journal of Obstetrics and Gynecology. 191, 1552–1572. - PubMed
-
- Bartling, B. , Desole, M. , Rohrbach, S. , Silber, R.E. , Simm, A. , 2009. Age-associated changes of extracellular matrix collagen impair lung cancer cell migration. Faseb Journal. 23, 1510–1520. - PubMed
-
- Barton, C.A. , Hacker, N.F. , Clark, S.J. , O'Brien, P.M. , 2008. DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecologic Oncology. 109, 129–139. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

