Pyrroloquinoline quinone biosynthesis gene pqqC, a novel molecular marker for studying the phylogeny and diversity of phosphate-solubilizing pseudomonads
- PMID: 21856827
- PMCID: PMC3194850
- DOI: 10.1128/AEM.05434-11
Pyrroloquinoline quinone biosynthesis gene pqqC, a novel molecular marker for studying the phylogeny and diversity of phosphate-solubilizing pseudomonads
Abstract
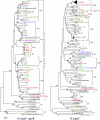
Many root-colonizing pseudomonads are able to promote plant growth by increasing phosphate availability in soil through solubilization of poorly soluble rock phosphates. The major mechanism of phosphate solubilization by pseudomonads is the secretion of gluconic acid, which requires the enzyme glucose dehydrogenase and its cofactor pyrroloquinoline quinone (PQQ). The main aim of this study was to evaluate whether a PQQ biosynthetic gene is suitable to study the phylogeny of phosphate-solubilizing pseudomonads. To this end, two new primers, which specifically amplify the pqqC gene of the Pseudomonas genus, were designed. pqqC fragments were amplified and sequenced from a Pseudomonas strain collection and from a natural wheat rhizosphere population using cultivation-dependent and cultivation-independent approaches. Phylogenetic trees based on pqqC sequences were compared to trees obtained with the two concatenated housekeeping genes rpoD and gyrB. For both pqqC and rpoD-gyrB, similar main phylogenetic clusters were found. However, in the pqqC but not in the rpoD-gyrB tree, the group of fluorescent pseudomonads producing the antifungal compounds 2,4-diacetylphloroglucinol and pyoluteorin was located outside the Pseudomonas fluorescens group. pqqC sequences from isolated pseudomonads were differently distributed among the identified phylogenetic groups than pqqC sequences derived from the cultivation-independent approach. Comparing pqqC phylogeny and phosphate solubilization activity, we identified one phylogenetic group with high solubilization activity. In summary, we demonstrate that the gene pqqC is a novel molecular marker that can be used complementary to housekeeping genes for studying the diversity and evolution of plant-beneficial pseudomonads.
Figures

References
-
- Brandl H., Lehmann S., Faramarzi M. A., Martinelli D. 2008. Biomobilization of silver, gold, and platinum from solid materials by HCN-forming microorganisms. Hydrometallurgy 94:14–17
-
- Browne P., et al. 2009. Superior inorganic phosphate solubilization is linked to phylogeny within the Pseudomonas fluorescens complex. Appl. Soil Ecol. 43:131–138
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

