Lateral habenula neurons signal errors in the prediction of reward information
- PMID: 21857659
- PMCID: PMC3164948
- DOI: 10.1038/nn.2902
Lateral habenula neurons signal errors in the prediction of reward information
Erratum in
- Nat Neurosci. 2011 Dec;14(12):1617
Abstract
Humans and animals have the ability to predict future events, which they cultivate by continuously searching their environment for sources of predictive information. However, little is known about the neural systems that motivate this behavior. We hypothesized that information-seeking is assigned value by the same circuits that support reward-seeking, such that neural signals encoding reward prediction errors (RPEs) include analogous information prediction errors (IPEs). To test this, we recorded from neurons in the lateral habenula, a nucleus that encodes RPEs, while monkeys chose between cues that provided different chances to view information about upcoming rewards. We found that a subpopulation of lateral habenula neurons transmitted signals resembling IPEs, responding when reward information was unexpectedly cued, delivered or denied. These signals evaluated information sources reliably, even when the monkey's decisions did not. These neurons could provide a common instructive signal for reward-seeking and information-seeking behavior.
Figures




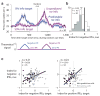
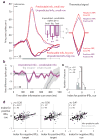


Comment in
-
On the value of information and other rewards.Nat Neurosci. 2011 Aug 26;14(9):1095-7. doi: 10.1038/nn.2918. Nat Neurosci. 2011. PMID: 21878921 No abstract available.
References
-
- Schultz W, Dayan P, Montague PR. A neural substrate of prediction and reward. Science. 1997;275:1593–9. - PubMed
-
- Rescorla RA, Wagner AR. In: Classical Conditioning II: Current Research and Theory. Black AH, Prokasy WF, editors. Appleton Century Crofts; New York, New York: 1972. pp. 64–99.
-
- Matsumoto M, Matsumoto K, Abe H, Tanaka K. Medial prefrontal cell activity signaling prediction errors of action values. Nat Neurosci. 2007;10:647–56. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

