Multiple ligand-specific conformations of the β2-adrenergic receptor
- PMID: 21857662
- PMCID: PMC3404607
- DOI: 10.1038/nchembio.634
Multiple ligand-specific conformations of the β2-adrenergic receptor
Abstract
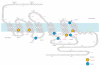
Seven-transmembrane receptors (7TMRs), also called G protein-coupled receptors (GPCRs), represent the largest class of drug targets, and they can signal through several distinct mechanisms including those mediated by G proteins and the multifunctional adaptor proteins β-arrestins. Moreover, several receptor ligands with differential efficacies toward these distinct signaling pathways have been identified. However, the structural basis and mechanism underlying this 'biased agonism' remains largely unknown. Here, we develop a quantitative mass spectrometry strategy that measures specific reactivities of individual side chains to investigate dynamic conformational changes in the β(2)-adrenergic receptor occupied by nine functionally distinct ligands. Unexpectedly, only a minority of residues showed reactivity patterns consistent with classical agonism, whereas the majority showed distinct patterns of reactivity even between functionally similar ligands. These findings demonstrate, contrary to two-state models for receptor activity, that there is significant variability in receptor conformations induced by different ligands, which has significant implications for the design of new therapeutic agents.
Figures


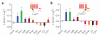
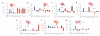
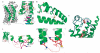
References
-
- Lefkowitz RJ. Seven transmembrane receptors: something old, something new. Acta Physiol. (Oxf.) 2007;190:9–19. - PubMed
-
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 2002;3:639–650. - PubMed
-
- Lagerström MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008;7:339–357. - PubMed
-
- Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. beta-Arrestin: a protein that regulates beta-adrenergic receptor function. Science. 1990;248:1547–1550. - PubMed
-
- Luttrell LM, Lefkowitz RJ. The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J. Cell Sci. 2002;115:455–465. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

