Statistical Mechanics of Nucleosomes Constrained by Higher-Order Chromatin Structure
- PMID: 21857746
- PMCID: PMC3156456
- DOI: 10.1007/s10955-011-0214-y
Statistical Mechanics of Nucleosomes Constrained by Higher-Order Chromatin Structure
Abstract
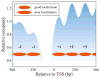
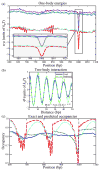
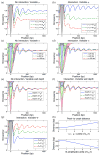
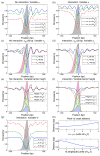
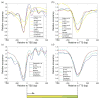
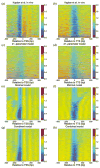
Eukaryotic DNA is packaged into chromatin: one-dimensional arrays of nucleosomes separated by stretches of linker DNA are folded into 30-nm chromatin fibers which in turn form higher-order structures (Felsenfeld and Groudine in Nature 421:448, 2003). Each nucleosome, the fundamental unit of chromatin, has 147 base pairs (bp) of DNA wrapped around a histone octamer (Richmond and Davey in Nature 423:145, 2003). In order to describe how chromatin fiber formation affects nucleosome positioning and energetics, we have developed a thermodynamic model of finite-size particles with effective nearest-neighbor interactions and arbitrary DNA-binding energies. We show that both one-and two-body interactions can be extracted from one-particle density profiles based on high-throughput maps of in vitro or in vivo nucleosome positions. Although a simpler approach that neglects two-body interactions (even if they are in fact present in the system) can be used to predict sequence determinants of nucleosome positions, the full theory is required to disentangle one- and two-body effects. Finally, we construct a minimal model in which nucleosomes are positioned primarily by steric exclusion and two-body interactions rather than intrinsic histone-DNA sequence preferences. The model reproduces nucleosome occupancy patterns observed over transcribed regions in living cells.
Figures
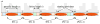



References
Grants and funding
LinkOut - more resources
Full Text Sources
