Real-time PyMOL visualization for Rosetta and PyRosetta
- PMID: 21857909
- PMCID: PMC3156697
- DOI: 10.1371/journal.pone.0021931
Real-time PyMOL visualization for Rosetta and PyRosetta
Abstract
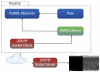
Computational structure prediction and design of proteins and protein-protein complexes have long been inaccessible to those not directly involved in the field. A key missing component has been the ability to visualize the progress of calculations to better understand them. Rosetta is one simulation suite that would benefit from a robust real-time visualization solution. Several tools exist for the sole purpose of visualizing biomolecules; one of the most popular tools, PyMOL (Schrödinger), is a powerful, highly extensible, user friendly, and attractive package. Integrating Rosetta and PyMOL directly has many technical and logistical obstacles inhibiting usage. To circumvent these issues, we developed a novel solution based on transmitting biomolecular structure and energy information via UDP sockets. Rosetta and PyMOL run as separate processes, thereby avoiding many technical obstacles while visualizing information on-demand in real-time. When Rosetta detects changes in the structure of a protein, new coordinates are sent over a UDP network socket to a PyMOL instance running a UDP socket listener. PyMOL then interprets and displays the molecule. This implementation also allows remote execution of Rosetta. When combined with PyRosetta, this visualization solution provides an interactive environment for protein structure prediction and design.
Conflict of interest statement
Figures
References
-
- Das R, Baker D. Macromolecular modeling with Rosetta. Biochemistry. 2008;77:363–382. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

