Evolutionary genomics implies a specific function of Ant4 in mammalian and anole lizard male germ cells
- PMID: 21858006
- PMCID: PMC3155547
- DOI: 10.1371/journal.pone.0023122
Evolutionary genomics implies a specific function of Ant4 in mammalian and anole lizard male germ cells
Abstract
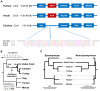
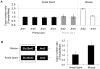
Most vertebrates have three paralogous genes with identical intron-exon structures and a high degree of sequence identity that encode mitochondrial adenine nucleotide translocase (Ant) proteins, Ant1 (Slc25a4), Ant2 (Slc25a5) and Ant3 (Slc25a6). Recently, we and others identified a fourth mammalian Ant paralog, Ant4 (Slc25a31), with a distinct intron-exon structure and a lower degree of sequence identity. Ant4 was expressed selectively in testis and sperm in adult mammals and was indeed essential for mouse spermatogenesis, but it was absent in birds, fish and frogs. Since Ant2 is X-linked in mammalian genomes, we hypothesized that the autosomal Ant4 gene may compensate for the loss of Ant2 gene expression during male meiosis in mammals. Here we report that the Ant4 ortholog is conserved in green anole lizard (Anolis carolinensis) and demonstrate that it is expressed in the anole testis. Further, a degenerate DNA fragment of putative Ant4 gene was identified in syntenic regions of avian genomes, indicating that Ant4 was present in the common amniote ancestor. Phylogenetic analyses suggest an even more ancient origin of the Ant4 gene. Although anole lizards are presumed male (XY) heterogametic, like mammals, copy numbers of the Ant2 as well as its neighboring gene were similar between male and female anole genomes, indicating that the anole Ant2 gene is either autosomal or located in the pseudoautosomal region of the sex chromosomes, in contrast to the case to mammals. These results imply the conservation of Ant4 is not likely simply driven by the sex chromosomal localization of the Ant2 gene and its subsequent inactivation during male meiosis. Taken together with the fact that Ant4 protein has a uniquely conserved structure when compared to other somatic Ant1, 2 and 3, there may be a specific advantage for mammals and lizards to express Ant4 in their male germ cells.
Conflict of interest statement
Figures




Similar articles
-
Evolutionarily conserved mammalian adenine nucleotide translocase 4 is essential for spermatogenesis.J Biol Chem. 2007 Oct 5;282(40):29658-66. doi: 10.1074/jbc.M704386200. Epub 2007 Aug 6. J Biol Chem. 2007. PMID: 17681941
-
Isolation, nucleotide identification and tissue expression of three novel ovine genes-SLC25A4, SLC25A5 and SLC25A6.Mol Biol Rep. 2010 Jul;37(6):2743-8. doi: 10.1007/s11033-009-9812-z. Epub 2009 Sep 11. Mol Biol Rep. 2010. PMID: 19763879
-
Adenine nucleotide translocase 4 is expressed within embryonic ovaries and dispensable during oogenesis.Reprod Sci. 2015 Feb;22(2):250-7. doi: 10.1177/1933719114542026. Epub 2014 Jul 16. Reprod Sci. 2015. PMID: 25031318 Free PMC article.
-
Adenine nucleotide translocase 2 is a key mitochondrial protein in cancer metabolism.Biochim Biophys Acta. 2011 Jun;1807(6):562-7. doi: 10.1016/j.bbabio.2010.10.008. Epub 2010 Oct 13. Biochim Biophys Acta. 2011. PMID: 20950584 Review.
-
The mitochondrial ADP/ATP carrier (SLC25 family): pathological implications of its dysfunction.Mol Aspects Med. 2013 Apr-Jun;34(2-3):485-93. doi: 10.1016/j.mam.2012.05.006. Mol Aspects Med. 2013. PMID: 23506884 Review.
Cited by
-
Mitochondrial Permeability Transition in Stem Cells, Development, and Disease.Adv Exp Med Biol. 2023;1409:1-22. doi: 10.1007/5584_2022_720. Adv Exp Med Biol. 2023. PMID: 35739412 Review.
-
Two adenine nucleotide translocase paralogues involved in cell proliferation and spermatogenesis in the silkworm Bombyx mori.PLoS One. 2015 Mar 5;10(3):e0119429. doi: 10.1371/journal.pone.0119429. eCollection 2015. PLoS One. 2015. PMID: 25742135 Free PMC article.
-
Sperm Differentiation: The Role of Trafficking of Proteins.Int J Mol Sci. 2020 May 24;21(10):3702. doi: 10.3390/ijms21103702. Int J Mol Sci. 2020. PMID: 32456358 Free PMC article. Review.
-
Genome reannotation of the lizard Anolis carolinensis based on 14 adult and embryonic deep transcriptomes.BMC Genomics. 2013 Jan 23;14:49. doi: 10.1186/1471-2164-14-49. BMC Genomics. 2013. PMID: 23343042 Free PMC article.
-
Inhibition of mitochondrial permeability transition by deletion of the ANT family and CypD.Sci Adv. 2019 Aug 28;5(8):eaaw4597. doi: 10.1126/sciadv.aaw4597. eCollection 2019 Aug. Sci Adv. 2019. PMID: 31489369 Free PMC article.
References
-
- Klingenberg M. Molecular aspects of the adenine nucleotide carrier from mitochondria. Arch Biochem Biophys. 1989;270:1–14. - PubMed
-
- Nelson DR, Felix CM, Swanson JM. Highly conserved charge-pair networks in the mitochondrial carrier family. J Mol Biol. 1998;277:285–308. - PubMed
-
- Fiore C, Trezeguet V, Le Saux A, Roux P, Schwimmer C, et al. The mitochondrial ADP/ATP carrier: structural, physiological and pathological aspects. Biochimie. 1998;80:137–150. - PubMed
-
- Palmieri L, Lasorsa FM, Vozza A, Agrimi G, Fiermonte G, et al. Identification and functions of new transporters in yeast mitochondria. Biochim Biophys Acta. 2000;1459:363–369. - PubMed
-
- Amiri H, Karlberg O, Andersson SG. Deep origin of plastid/parasite ATP/ADP translocases. J Mol Evol. 2003;56:137–150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

