HIV protein sequence hotspots for crosstalk with host hub proteins
- PMID: 21858059
- PMCID: PMC3156123
- DOI: 10.1371/journal.pone.0023293
HIV protein sequence hotspots for crosstalk with host hub proteins
Abstract
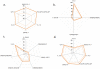
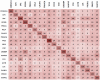
HIV proteins target host hub proteins for transient binding interactions. The presence of viral proteins in the infected cell results in out-competition of host proteins in their interaction with hub proteins, drastically affecting cell physiology. Functional genomics and interactome datasets can be used to quantify the sequence hotspots on the HIV proteome mediating interactions with host hub proteins. In this study, we used the HIV and human interactome databases to identify HIV targeted host hub proteins and their host binding partners (H2). We developed a high throughput computational procedure utilizing motif discovery algorithms on sets of protein sequences, including sequences of HIV and H2 proteins. We identified as HIV sequence hotspots those linear motifs that are highly conserved on HIV sequences and at the same time have a statistically enriched presence on the sequences of H2 proteins. The HIV protein motifs discovered in this study are expressed by subsets of H2 host proteins potentially outcompeted by HIV proteins. A large subset of these motifs is involved in cleavage, nuclear localization, phosphorylation, and transcription factor binding events. Many such motifs are clustered on an HIV sequence in the form of hotspots. The sequential positions of these hotspots are consistent with the curated literature on phenotype altering residue mutations, as well as with existing binding site data. The hotspot map produced in this study is the first global portrayal of HIV motifs involved in altering the host protein network at highly connected hub nodes.
Conflict of interest statement
Figures

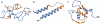
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

