Epigenetic transcriptional regulation of the growth arrest-specific gene 1 (Gas1) in hepatic cell proliferation at mononucleosomal resolution
- PMID: 21858068
- PMCID: PMC3153484
- DOI: 10.1371/journal.pone.0023318
Epigenetic transcriptional regulation of the growth arrest-specific gene 1 (Gas1) in hepatic cell proliferation at mononucleosomal resolution
Abstract
Background: Gas1 (growth arrest-specific 1) gene is known to inhibit cell proliferation in a variety of models, but its possible implication in regulating quiescence in adult tissues has not been examined to date. The knowledge of how Gas1 is regulated in quiescence may contribute to understand the deregulation occurring in neoplastic diseases.
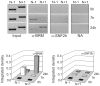
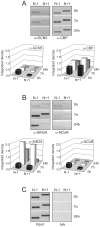
Methodology/principal findings: Gas1 expression has been studied in quiescent murine liver and during the naturally synchronized cell proliferation after partial hepatectomy. Chromatin immunoprecipitation at nucleosomal resolution (Nuc-ChIP) has been used to carry out the study preserving the in vivo conditions. Transcription has been assessed at real time by quantifying the presence of RNA polymerase II in coding regions (RNApol-ChIP). It has been found that Gas1 is expressed not only in quiescent liver but also at the cell cycle G(1)/S transition. The latter expression peak had not been previously reported. Two nucleosomes, flanking a nucleosome-free region, are positioned close to the transcription start site. Both nucleosomes slide in going from the active to the inactive state and vice versa. Nuc-ChIP analysis of the acquisition of histone epigenetic marks show distinctive features in both active states: H3K9ac and H3K4me2 are characteristic of transcription in G(0) and H4R3me2 in G(1)/S transition. Sequential-ChIP analysis revealed that the "repressing" mark H3K9me2 colocalize with several "activating" marks at nucleosome N-1 when Gas1 is actively transcribed suggesting a greater plasticity of epigenetic marks than proposed until now. The recruitment of chromatin-remodeling or modifying complexes also displayed distinct characteristics in quiescence and the G(1)/S transition.
Conclusions/significance: The finding that Gas1 is transcribed at the G(1)/S transition suggests that the gene may exert a novel function during cell proliferation. Transcription of this gene is modulated by specific "activating" and "repressing" epigenetic marks, and by chromatin remodeling and histone modifying complexes recruitment, at specific nucleosomes in Gas1 promoter.
Conflict of interest statement
Figures



References
-
- Fausto N. Liver regeneration. J Hepatol. 2000;32:19–31. - PubMed
-
- Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45–53. - PubMed
-
- Fausto N, Laird AD, Webber EM. Liver regeneration. 2. Role of growth factors, cytokines in hepatic regeneration. FASEB J. 1995;9:1527–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

