A cytoplasmic negative regulator isoform of ATF7 impairs ATF7 and ATF2 phosphorylation and transcriptional activity
- PMID: 21858082
- PMCID: PMC3156760
- DOI: 10.1371/journal.pone.0023351
A cytoplasmic negative regulator isoform of ATF7 impairs ATF7 and ATF2 phosphorylation and transcriptional activity
Abstract
Alternative splicing and post-translational modifications are processes that give rise to the complexity of the proteome. The nuclear ATF7 and ATF2 (activating transcription factor) are structurally homologous leucine zipper transcription factors encoded by distinct genes. Stress and growth factors activate ATF2 and ATF7 mainly via sequential phosphorylation of two conserved threonine residues in their activation domain. Distinct protein kinases, among which mitogen-activated protein kinases (MAPK), phosphorylate ATF2 and ATF7 first on Thr71/Thr53 and next on Thr69/Thr51 residues respectively, resulting in transcriptional activation. Here, we identify and characterize a cytoplasmic alternatively spliced isoform of ATF7. This variant, named ATF7-4, inhibits both ATF2 and ATF7 transcriptional activities by impairing the first phosphorylation event on Thr71/Thr53 residues. ATF7-4 indeed sequesters the Thr53-phosphorylating kinase in the cytoplasm. Upon stimulus-induced phosphorylation, ATF7-4 is poly-ubiquitinated and degraded, enabling the release of the kinase and ATF7/ATF2 activation. Our data therefore conclusively establish that ATF7-4 is an important cytoplasmic negative regulator of ATF7 and ATF2 transcription factors.
Conflict of interest statement
Figures


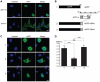

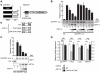

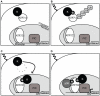
Similar articles
-
p38beta2-mediated phosphorylation and sumoylation of ATF7 are mutually exclusive.J Mol Biol. 2008 Dec 26;384(4):980-91. doi: 10.1016/j.jmb.2008.10.008. Epub 2008 Oct 11. J Mol Biol. 2008. PMID: 18950637
-
Cdk1-mediated phosphorylation of human ATF7 at Thr-51 and Thr-53 promotes cell-cycle progression into M phase.PLoS One. 2014 Dec 29;9(12):e116048. doi: 10.1371/journal.pone.0116048. eCollection 2014. PLoS One. 2014. PMID: 25545367 Free PMC article.
-
ATF2 and ATF7 Are Critical Mediators of Intestinal Epithelial Repair.Cell Mol Gastroenterol Hepatol. 2020;10(1):23-42. doi: 10.1016/j.jcmgh.2020.01.005. Epub 2020 Jan 17. Cell Mol Gastroenterol Hepatol. 2020. PMID: 31958521 Free PMC article.
-
The roles of ATF2 (activating transcription factor 2) in tumorigenesis.Biochem Soc Trans. 2012 Feb;40(1):230-4. doi: 10.1042/BST20110630. Biochem Soc Trans. 2012. PMID: 22260696 Review.
-
ATF2 on the double - activating transcription factor and DNA damage response protein.Pigment Cell Res. 2007 Dec;20(6):498-506. doi: 10.1111/j.1600-0749.2007.00414.x. Pigment Cell Res. 2007. PMID: 17935492 Free PMC article. Review.
Cited by
-
The activating transcription factor 2: an influencer of cancer progression.Mutagenesis. 2019 Dec 19;34(5-6):375-389. doi: 10.1093/mutage/gez041. Mutagenesis. 2019. PMID: 31799611 Free PMC article. Review.
-
Systematic Analysis of the Genetic Variability That Impacts SUMO Conjugation and Their Involvement in Human Diseases.Sci Rep. 2015 Jul 8;5:10900. doi: 10.1038/srep10900. Sci Rep. 2015. PMID: 26154679 Free PMC article.
-
The two-faced role of ATF2 on cisplatin response in gastric cancer depends on p53 context.Cell Biosci. 2022 May 31;12(1):77. doi: 10.1186/s13578-022-00802-w. Cell Biosci. 2022. PMID: 35641966 Free PMC article.
-
ATF7 is stabilized during mitosis in a CDK1-dependent manner and contributes to cyclin D1 expression.Cell Cycle. 2015;14(16):2655-66. doi: 10.1080/15384101.2015.1064568. Cell Cycle. 2015. PMID: 26101806 Free PMC article.
-
Exosomal MicroRNAs in Pregnancy Provides Insight into a Possible Cure for Cancer.Int J Mol Sci. 2020 Jul 29;21(15):5384. doi: 10.3390/ijms21155384. Int J Mol Sci. 2020. PMID: 32751127 Free PMC article.
References
-
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. - PubMed
-
- Nomura N, Zu YL, Maekawa T, Tabata S, Akiyama T, et al. Isolation and characterization of a novel member of the gene family encoding the cAMP response element-binding protein CRE-BP1. JBiolChem. 1993;268:4259–4266. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

