High diversity of the saliva microbiome in Batwa Pygmies
- PMID: 21858083
- PMCID: PMC3156759
- DOI: 10.1371/journal.pone.0023352
High diversity of the saliva microbiome in Batwa Pygmies
Abstract
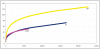
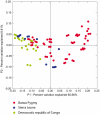
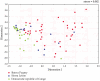
We describe the saliva microbiome diversity in Batwa Pygmies, a former hunter-gatherer group from Uganda, using next-generation sequencing of partial 16S rRNA sequences. Microbial community diversity in the Batwa is significantly higher than in agricultural groups from Sierra Leone and the Democratic Republic of Congo. We found 40 microbial genera in the Batwa, which have previously not been described in the human oral cavity. The distinctive composition of the salvia microbiome of the Batwa may have been influenced by their recent different lifestyle and diet.
Conflict of interest statement
Figures


References
-
- Gusinde M. Pygmies and Pygmoids: Twides of tropical Africa. Anthropological Quarterly. 1955;28:3–61.
-
- Walker PL, Hewlett BS. Dental Health Diet and Social Status among Central African Foragers and Farmers. American Anthropologist, New series. 1990;92:383–398.
-
- Quinque D, Kittler R, Kayser M, Stoneking M, Nasidze I. Evaluation of saliva as a source of human DNA for population and association studies. Analytical Biochemistry. 2006;353:272–277. - PubMed
-
- Meyer M, Stenzel U, Hofreiter M. Parallel tagged sequencing on the 454 platform. Nat Protoc. 2008;3:267–278. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

