Unique changes in mitochondrial genomes associated with reversions of S-type cytoplasmic male sterility in maizemar
- PMID: 21858103
- PMCID: PMC3152571
- DOI: 10.1371/journal.pone.0023405
Unique changes in mitochondrial genomes associated with reversions of S-type cytoplasmic male sterility in maizemar
Abstract
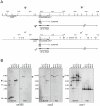
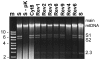
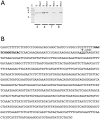
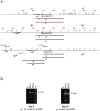
Cytoplasmic male sterility (CMS) in plants is usually associated with the expression of specific chimeric regions within rearranged mitochondrial genomes. Maize CMS-S plants express high amounts of a 1.6-kb mitochondrial RNA during microspore maturation, which is associated with the observed pollen abortion. This transcript carries two chimeric open reading frames, orf355 and orf77, both unique to CMS-S. CMS-S mitochondria also contain free linear DNA plasmids bearing terminal inverted repeats (TIRs). These TIRs recombine with TIR-homologous sequences that precede orf355/orf77 within the main mitochondrial genome to produce linear ends. Transcription of the 1.6-kb RNA is initiated from a promoter within the TIRs only when they are at linear ends. Reversions of CMS-S to fertility occur in certain nuclear backgrounds and are usually associated with loss of the S plasmids and/or the sterility-associated region. We describe an unusual set of independently recovered revertants from a single maternal lineage that retain both the S plasmids and an intact orf355/orf77 region but which do not produce the 1.6-kb RNA. A 7.3-kb inversion resulting from illegitimate recombination between 14-bp microrepeats has separated the genomic TIR sequences from the CMS-associated region. Although RNAs containing orf355/orf77 can still be detected in the revertants, they are not highly expressed during pollen development and they are no longer initiated from the TIR promoter at a protein-stabilized linear end. They appear instead to be co-transcribed with cytochrome oxidase subunit 2. The 7.3-kb inversion was not detected in CMS-S or in other fertile revertants. Therefore, this inversion appears to be a de novo mutation that has continued to sort out within a single maternal lineage, giving rise to fertile progeny in successive generations.
Conflict of interest statement
Figures


Similar articles
-
Expression analysis of orf77 and R region in mitochondrial DNA of S-type CMS maize.Yi Chuan Xue Bao. 2003 Mar;30(3):277-82. Yi Chuan Xue Bao. 2003. PMID: 12812094
-
Comparative analysis of mitochondrial genomes of maize CMS-S subtypes provides new insights into male sterility stability.BMC Plant Biol. 2022 Oct 1;22(1):469. doi: 10.1186/s12870-022-03849-6. BMC Plant Biol. 2022. PMID: 36180833 Free PMC article.
-
Characterization of a novel thermosensitive restorer of fertility for cytoplasmic male sterility in maize.Genetics. 2009 May;182(1):91-103. doi: 10.1534/genetics.108.099895. Epub 2009 Mar 2. Genetics. 2009. PMID: 19255365 Free PMC article.
-
Genome barriers between nuclei and mitochondria exemplified by cytoplasmic male sterility.Plant Cell Physiol. 2008 Oct;49(10):1484-94. doi: 10.1093/pcp/pcn102. Epub 2008 Jul 14. Plant Cell Physiol. 2008. PMID: 18625609 Free PMC article. Review.
-
Plant mitochondrial mutations and male sterility.Annu Rev Genet. 1991;25:461-86. doi: 10.1146/annurev.ge.25.120191.002333. Annu Rev Genet. 1991. PMID: 1812814 Review.
Cited by
-
Mitochondrial Transcriptome Control and Intercompartment Cross-Talk During Plant Development.Cells. 2019 Jun 13;8(6):583. doi: 10.3390/cells8060583. Cells. 2019. PMID: 31200566 Free PMC article.
-
The comparison of four mitochondrial genomes reveals cytoplasmic male sterility candidate genes in cotton.BMC Genomics. 2018 Oct 26;19(1):775. doi: 10.1186/s12864-018-5122-y. BMC Genomics. 2018. PMID: 30367630 Free PMC article.
-
Identification of Genes Potentially Associated with the Fertility Instability of S-Type Cytoplasmic Male Sterility in Maize via Bulked Segregant RNA-Seq.PLoS One. 2016 Sep 26;11(9):e0163489. doi: 10.1371/journal.pone.0163489. eCollection 2016. PLoS One. 2016. PMID: 27669430 Free PMC article.
-
Comparative analysis of mitochondrial genomes of soybean cytoplasmic male-sterile lines and their maintainer lines.Funct Integr Genomics. 2021 Jan;21(1):43-57. doi: 10.1007/s10142-020-00760-x. Epub 2021 Jan 6. Funct Integr Genomics. 2021. PMID: 33404916
-
Identification of fertility-related genes for maize CMS-S via Bulked Segregant RNA-Seq.PeerJ. 2020 Sep 30;8:e10015. doi: 10.7717/peerj.10015. eCollection 2020. PeerJ. 2020. PMID: 33062436 Free PMC article.
References
-
- Fauron C, Casper M, Gao Y, Moore B. The maize mitochondrial genome: dynamic, yet functional. Trends Genet. 1995;11:228–235. - PubMed
-
- Newton KJ, Gabay-Laughnan S, DePaepe R. D.A. Day HA, Millar, Whelan J, editors. Mitochondrial mutations in plants. Plant Mitochondria: From Genome to Function. 2004. pp. 121–142.
-
- Kubo T, Newton KJ. Angiosperm mitochondrial genomes and mutations. Mitochondrion. 2008;8:5–14. - PubMed
-
- Schardl CL, Lonsdale DM, Pring DR, Rose KR. Linearization of maize mitochondrial chromosomes by recombination with linear episomes. Nature. 1984;310:292–296.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

