The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life
- PMID: 21858139
- PMCID: PMC3153485
- DOI: 10.1371/journal.pone.0023479
The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life
Abstract
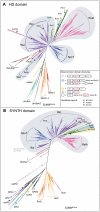
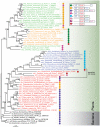
RelA/SpoT Homologue (RSH) proteins, named for their sequence similarity to the RelA and SpoT enzymes of Escherichia coli, comprise a superfamily of enzymes that synthesize and/or hydrolyze the alarmone ppGpp, activator of the "stringent" response and regulator of cellular metabolism. The classical "long" RSHs Rel, RelA and SpoT with the ppGpp hydrolase, synthetase, TGS and ACT domain architecture have been found across diverse bacteria and plant chloroplasts, while dedicated single domain ppGpp-synthesizing and -hydrolyzing RSHs have also been discovered in disparate bacteria and animals respectively. However, there is considerable confusion in terms of nomenclature and no comprehensive phylogenetic and sequence analyses have previously been carried out to classify RSHs on a genomic scale. We have performed high-throughput sensitive sequence searching of over 1000 genomes from across the tree of life, in combination with phylogenetic analyses to consolidate previous ad hoc identification of diverse RSHs in different organisms and provide a much-needed unifying terminology for the field. We classify RSHs into 30 subgroups comprising three groups: long RSHs, small alarmone synthetases (SASs), and small alarmone hydrolases (SAHs). Members of nineteen previously unidentified RSH subgroups can now be studied experimentally, including previously unknown RSHs in archaea, expanding the "stringent response" to this domain of life. We have analyzed possible combinations of RSH proteins and their domains in bacterial genomes and compared RSH content with available RSH knock-out data for various organisms to determine the rules of combining RSHs. Through comparative sequence analysis of long and small RSHs, we find exposed sites limited in conservation to the long RSHs that we propose are involved in transmitting regulatory signals. Such signals may be transmitted via NTD to CTD intra-molecular interactions, or inter-molecular interactions either among individual RSH molecules or among long RSHs and other binding partners such as the ribosome.
Conflict of interest statement
Figures




References
-
- Pesavento C, Hengge R. Bacterial nucleotide-based second messengers. Curr Opin Microbiol. 2009;12:170–176. - PubMed
-
- Mittenhuber G. Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J Mol Microbiol Biotechnol. 2001;3:585–600. - PubMed
-
- Gallant J, Palmer L, Pao CC. Anomalous synthesis of ppGpp in growing cells. Cell. 1977;11:181–185. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

