c-Myc regulates self-renewal in bronchoalveolar stem cells
- PMID: 21858211
- PMCID: PMC3157444
- DOI: 10.1371/journal.pone.0023707
c-Myc regulates self-renewal in bronchoalveolar stem cells
Abstract
Background: Bronchoalveolar stem cells (BASCs) located in the bronchoalveolar duct junction are thought to regenerate both bronchiolar and alveolar epithelium during homeostatic turnover and in response to injury. The mechanisms directing self-renewal in BASCs are poorly understood.
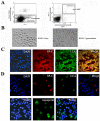
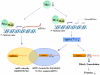
Methods: BASCs (Sca-1(+), CD34(+), CD31(-) and, CD45(-)) were isolated from adult mouse lung using FACS, and their capacity for self-renewal and differentiation were demonstrated by immunostaining. A transcription factor network of 53 genes required for pluripotency in embryonic stem cells was assessed in BASCs, Kras-initiated lung tumor tissue, and lung organogenesis by real-time PCR. c-Myc was knocked down in BASCs by infection with c-Myc shRNA lentivirus. Comprehensive miRNA and mRNA profiling for BASCs was performed, and significant miRNAs and mRNAs potentially regulated by c-Myc were identified. We explored a c-Myc regulatory network in BASCs using a number of statistical and computational approaches through two different strategies; 1) c-Myc/Max binding sites within individual gene promoters, and 2) miRNA-regulated target genes.
Results: c-Myc expression was upregulated in BASCs and downregulated over the time course of lung organogenesis in vivo. The depletion of c-Myc in BASCs resulted in decreased proliferation and cell death. Multiple mRNAs and miRNAs were dynamically regulated in c-Myc depleted BASCs. Among a total of 250 dynamically regulated genes in c-Myc depleted BASCs, 57 genes were identified as potential targets of miRNAs through miRBase and TargetScan-based computational mapping. A further 88 genes were identified as potential downstream targets through their c-Myc binding motif.
Conclusion: c-Myc plays a critical role in maintaining the self-renewal capacity of lung bronchoalveolar stem cells through a combination of miRNA and transcription factor regulatory networks.
Conflict of interest statement
Figures




Similar articles
-
MicroRNA expression profile of bronchioalveolar stem cells from mouse lung.Biochem Biophys Res Commun. 2008 Dec 12;377(2):668-673. doi: 10.1016/j.bbrc.2008.10.052. Epub 2008 Oct 21. Biochem Biophys Res Commun. 2008. PMID: 18948085
-
Myc and max genome-wide binding sites analysis links the Myc regulatory network with the polycomb and the core pluripotency networks in mouse embryonic stem cells.PLoS One. 2014 Feb 21;9(2):e88933. doi: 10.1371/journal.pone.0088933. eCollection 2014. PLoS One. 2014. PMID: 24586446 Free PMC article.
-
myc maintains embryonic stem cell pluripotency and self-renewal.Differentiation. 2010 Jul;80(1):9-19. doi: 10.1016/j.diff.2010.05.001. Epub 2010 May 27. Differentiation. 2010. PMID: 20537458 Free PMC article.
-
Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells?Curr Stem Cell Res Ther. 2009 Sep;4(3):168-77. doi: 10.2174/157488809789057400. Curr Stem Cell Res Ther. 2009. PMID: 19492978 Review.
-
Bronchioalveolar stem cells in lung repair, regeneration and disease.J Pathol. 2020 Nov;252(3):219-226. doi: 10.1002/path.5527. Epub 2020 Oct 1. J Pathol. 2020. PMID: 32737996 Review.
Cited by
-
Control of vertebrate development by MYC.Cold Spring Harb Perspect Med. 2013 Sep 1;3(9):a014332. doi: 10.1101/cshperspect.a014332. Cold Spring Harb Perspect Med. 2013. PMID: 24003246 Free PMC article. Review.
-
Myc Cooperates with Ras by Programming Inflammation and Immune Suppression.Cell. 2017 Nov 30;171(6):1301-1315.e14. doi: 10.1016/j.cell.2017.11.013. Cell. 2017. PMID: 29195074 Free PMC article.
-
cMyc Regulates the Size of the Premigratory Neural Crest Stem Cell Pool.Cell Rep. 2016 Dec 6;17(10):2648-2659. doi: 10.1016/j.celrep.2016.11.025. Cell Rep. 2016. PMID: 27926868 Free PMC article.
-
Unraveling MYC's Role in Orchestrating Tumor Intrinsic and Tumor Microenvironment Interactions Driving Tumorigenesis and Drug Resistance.Pathophysiology. 2023 Sep 11;30(3):400-419. doi: 10.3390/pathophysiology30030031. Pathophysiology. 2023. PMID: 37755397 Free PMC article. Review.
-
MicroRNA-mRNA interactions in a murine model of hyperoxia-induced bronchopulmonary dysplasia.BMC Genomics. 2012 May 30;13:204. doi: 10.1186/1471-2164-13-204. BMC Genomics. 2012. PMID: 22646479 Free PMC article.
References
-
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. - PubMed
-
- Qian S, Ding JY, Xie R, An JH, Ao XJ, et al. MicroRNA expression profile of bronchioalveolar stem cells from mouse lung. Biochem Biophys Res Commun. 2008;377:668–673. - PubMed
-
- Wuenschell CW, Sunday ME, Singh G, Minoo P, Slavkin HC, et al. Embryonic mouse lung epithelial progenitor cells co-express immunohistochemical markers of diverse mature cell lineages. J Histochem Cytochem. 1996;44:113–123. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

