Identification of a novel ZIC3 isoform and mutation screening in patients with heterotaxy and congenital heart disease
- PMID: 21858219
- PMCID: PMC3157443
- DOI: 10.1371/journal.pone.0023755
Identification of a novel ZIC3 isoform and mutation screening in patients with heterotaxy and congenital heart disease
Abstract
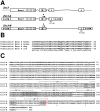
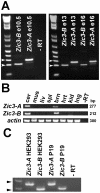
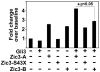
Patients with heterotaxy have characteristic cardiovascular malformations, abnormal arrangement of their visceral organs, and midline patterning defects that result from abnormal left-right patterning during embryogenesis. Loss of function of the transcription factor ZIC3 causes X-linked heterotaxy and isolated congenital heart malformations and represents one of the few known monogenic causes of congenital heart disease. The birth incidence of heterotaxy-spectrum malformations is significantly higher in males, but our previous work indicated that mutations within ZIC3 did not account for the male over-representation. Therefore, cross species comparative sequence alignment was used to identify a putative novel fourth exon, and the existence of a novel alternatively spliced transcript was confirmed by amplification from murine embryonic RNA and subsequent sequencing. This transcript, termed Zic3-B, encompasses exons 1, 2, and 4 whereas Zic3-A encompasses exons 1, 2, and 3. The resulting protein isoforms are 466 and 456 amino acid residues respectively, sharing the first 407 residues. Importantly, the last two amino acids in the fifth zinc finger DNA binding domain are altered in the Zic3-B isoform, indicating a potential functional difference that was further evaluated by expression, subcellular localization, and transactivation analyses. The temporo-spatial expression pattern of Zic3-B overlaps with Zic3-A in vivo, and both isoforms are localized to the nucleus in vitro. Both isoforms can transcriptionally activate a Gli binding site reporter, but only ZIC3-A synergistically activates upon co-transfection with Gli3, suggesting that the isoforms are functionally distinct. Screening 109 familial and sporadic male heterotaxy cases did not identify pathogenic mutations in the newly identified fourth exon and larger studies are necessary to establish the importance of the novel isoform in human disease.
Conflict of interest statement
Figures




Similar articles
-
Rare novel variants in the ZIC3 gene cause X-linked heterotaxy.Eur J Hum Genet. 2016 Dec;24(12):1783-1791. doi: 10.1038/ejhg.2016.91. Epub 2016 Jul 13. Eur J Hum Genet. 2016. PMID: 27406248 Free PMC article.
-
Identification and functional analysis of ZIC3 mutations in heterotaxy and related congenital heart defects.Am J Hum Genet. 2004 Jan;74(1):93-105. doi: 10.1086/380998. Epub 2003 Dec 16. Am J Hum Genet. 2004. PMID: 14681828 Free PMC article.
-
Non-coding cause of congenital heart defects: Abnormal RNA splicing with multiple isoforms as a mechanism for heterotaxy.HGG Adv. 2024 Oct 10;5(4):100353. doi: 10.1016/j.xhgg.2024.100353. Epub 2024 Sep 12. HGG Adv. 2024. PMID: 39275801 Free PMC article.
-
ZIC3 in Heterotaxy.Adv Exp Med Biol. 2018;1046:301-327. doi: 10.1007/978-981-10-7311-3_15. Adv Exp Med Biol. 2018. PMID: 29442328 Free PMC article. Review.
-
The role of ZIC3 in vertebrate development.Cytogenet Genome Res. 2002;99(1-4):229-35. doi: 10.1159/000071598. Cytogenet Genome Res. 2002. PMID: 12900569 Review.
Cited by
-
Splenic Torsion in Heterotaxy Syndrome with Left Isomerism: A Case Report and Literature Review.Diagnostics (Basel). 2022 Nov 23;12(12):2920. doi: 10.3390/diagnostics12122920. Diagnostics (Basel). 2022. PMID: 36552927 Free PMC article.
-
Heterotaxy-spectrum heart defects in Zic3 hypomorphic mice.Pediatr Res. 2013 Nov;74(5):494-502. doi: 10.1038/pr.2013.147. Epub 2013 Sep 2. Pediatr Res. 2013. PMID: 23999067 Free PMC article.
-
Left Isomerism With Normal Bronchopulmonary Anatomy: Broadening the Heterotaxy Spectrum.Case Rep Radiol. 2025 Apr 17;2025:5512404. doi: 10.1155/crra/5512404. eCollection 2025. Case Rep Radiol. 2025. PMID: 40276594 Free PMC article.
-
Posttranscriptional Regulation by Proteins and Noncoding RNAs.Adv Exp Med Biol. 2024;1441:313-339. doi: 10.1007/978-3-031-44087-8_17. Adv Exp Med Biol. 2024. PMID: 38884719
-
The determination factors of left-right asymmetry disorders- a short review.Clujul Med. 2017;90(2):139-146. doi: 10.15386/cjmed-701. Epub 2017 Apr 25. Clujul Med. 2017. PMID: 28559696 Free PMC article. Review.
References
-
- Clark KL, Yutzey KE, Benson DW. Transcription factors and congenital heart defects. Annu Rev Physiol. 2006;68:97–121. - PubMed
-
- Purandare SM, Ware SM, Kwan KM, Gebbia M, Bassi MT, et al. A complex syndrome of left-right axis, central nervous system and axial skeleton defects in Zic3 mutant mice. Development. 2002;129:2293–2302. - PubMed
-
- Zhu L, Zhou G, Poole S, Belmont JW. Characterization of the Interactions of Human ZIC3 Mutants With GLI3. Hum Mut. 2007;0:1–7. - PubMed
-
- Aruga J, Yokota N, Hashimoto M, Furuichi T, Fukuda M, et al. A novel zinc finger protein, zic, is involved in neurogenesis, especially in the cell lineage of cerebellar granule cells. J Neurochem. 1994;63:1880–1890. - PubMed
-
- Pavletich NP, Pabo CO. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993;261:1701–1707. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

