Matrin 3 binds and stabilizes mRNA
- PMID: 21858232
- PMCID: PMC3157474
- DOI: 10.1371/journal.pone.0023882
Matrin 3 binds and stabilizes mRNA
Abstract
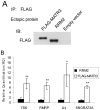
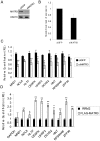
Matrin 3 (MATR3) is a highly conserved, inner nuclear matrix protein with two zinc finger domains and two RNA recognition motifs (RRM), whose function is largely unknown. Recently we found MATR3 to be phosphorylated by the protein kinase ATM, which activates the cellular response to double strand breaks in the DNA. Here, we show that MATR3 interacts in an RNA-dependent manner with several proteins with established roles in RNA processing, and maintains its interaction with RNA via its RRM2 domain. Deep sequencing of the bound RNA (RIP-seq) identified several small noncoding RNA species. Using microarray analysis to explore MATR3's role in transcription, we identified 77 transcripts whose amounts depended on the presence of MATR3. We validated this finding with nine transcripts which were also bound to the MATR3 complex. Finally, we demonstrated the importance of MATR3 for maintaining the stability of several of these mRNA species and conclude that it has a role in mRNA stabilization. The data suggest that the cellular level of MATR3, known to be highly regulated, modulates the stability of a group of gene transcripts.
Conflict of interest statement
Figures



References
-
- Belgrader P, Dey R, Berezney R. Molecular cloning of matrin 3. A 125-kilodalton protein of the nuclear matrix contains an extensive acidic domain. J Biol Chem. 1991;266:9893–9899. - PubMed
-
- Cohen TV, Hernandez L, Stewart CL. Functions of the nuclear envelope and lamina in development and disease. Biochem Soc Trans. 2008;36:1329–1334. - PubMed
-
- Hisada-Ishii S, Ebihara M, Kobayashi N, Kitagawa Y. Bipartite nuclear localization signal of matrin 3 is essential for vertebrate cells. Biochem Biophys Res Commun. 2007;354:72–76. - PubMed
-
- Hibino Y, Ohzeki H, Sugano N, Hiraga K. Transcription modulation by a rat nuclear scaffold protein, P130, and a rat highly repetitive DNA component or various types of animal and plant matrix or scaffold attachment regions. Biochem Biophys Res Commun. 2000;279:282–287. - PubMed
-
- Hibino Y, Usui T, Morita Y, Hirose N, Okazaki M, et al. Molecular properties and intracellular localization of rat liver nuclear scaffold protein P130. Biochim Biophys Acta. 2006;1759:195–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

