Comparative analysis of chromosome counts infers three paleopolyploidies in the mollusca
- PMID: 21859805
- PMCID: PMC3194838
- DOI: 10.1093/gbe/evr087
Comparative analysis of chromosome counts infers three paleopolyploidies in the mollusca
Abstract
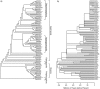
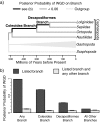
The study of paleopolyploidies requires the comparison of multiple whole genome sequences. If the branches of a phylogeny on which a whole-genome duplication (WGD) occurred could be identified before genome sequencing, taxa could be selected that provided a better assessment of that genome duplication. Here, we describe a likelihood model in which the number of chromosomes in a genome evolves according to a Markov process with one rate of chromosome duplication and loss that is proportional to the number of chromosomes in the genome and another stochastic rate at which every chromosome in the genome could duplicate in a single event. We compare the maximum likelihoods of a model in which the genome duplication rate varies to one in which it is fixed at zero using the Akaike information criterion, to determine if a model with WGDs is a good fit for the data. Once it has been determined that the data does fit the WGD model, we infer the phylogenetic position of paleopolyploidies by calculating the posterior probability that a WGD occurred on each branch of the taxon tree. Here, we apply this model to a molluscan tree represented by 124 taxa and infer three putative WGD events. In the Gastropoda, we identify a single branch within the Hypsogastropoda and one of two branches at the base of the Stylommatophora. We also identify one or two branches near the base of the Cephalopoda.
Figures




References
-
- Abi-Rached L, Gilles A, Shiina T, Pontarotti P, Inoko H. Evidence of en bloc duplication in vertebrate genomes. Nat Genet. 2002;31(1):100–105. - PubMed
-
- Ahmed M. Chromosome cytology of marine pelecypod molluscs. J Sci (Karachi). 1976;4:77–94.
-
- Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19(6):716–723.
-
- Anisimov AP, Tokmakova NP, Poveshchenko OS. Somatic polyploidy in some tissues of the succineid snail Succinea lauta (Gastropoda: Pulmonata) Tsitologiya. 1995;37(4):311–330.
-
- Ansorge WJ. Next-generation DNA sequencing techniques. N Biotechnol. 2009;25(4):195–203. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

