Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability
- PMID: 21859863
- PMCID: PMC3160583
- DOI: 10.1083/jcb.201102018
Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability
Abstract
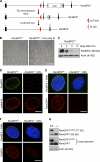
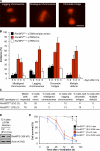
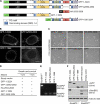
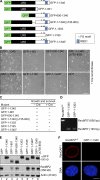
RanBP2/Nup358, the major component of the cytoplasmic filaments of the nuclear pore complex (NPC), is essential for mouse embryogenesis and is implicated in both macromolecular transport and mitosis, but its specific molecular functions are unknown. Using RanBP2 conditional knockout mouse embryonic fibroblasts and a series of mutant constructs, we show that transport, rather than mitotic, functions of RanBP2 are required for cell viability. Cre-mediated RanBP2 inactivation caused cell death with defects in M9- and classical nuclear localization signal (cNLS)-mediated protein import, nuclear export signal-mediated protein export, and messenger ribonucleic acid export but no apparent mitotic failure. A short N-terminal RanBP2 fragment harboring the NPC-binding domain, three phenylalanine-glycine motifs, and one Ran-binding domain (RBD) corrected all transport defects and restored viability. Mutation of the RBD within this fragment caused lethality and perturbed binding to Ran guanosine triphosphate (GTP)-importin-β, accumulation of importin-β at nuclear pores, and cNLS-mediated protein import. These data suggest that a critical function of RanBP2 is to capture recycling RanGTP-importin-β complexes at cytoplasmic fibrils to allow for adequate cNLS-mediated cargo import.
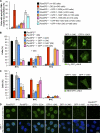
Figures



References
-
- Aslanukov A., Bhowmick R., Guruju M., Oswald J., Raz D., Bush R.A., Sieving P.A., Lu X., Bock C.B., Ferreira P.A. 2006. RanBP2 modulates Cox11 and hexokinase I activities and haploinsufficiency of RanBP2 causes deficits in glucose metabolism. PLoS Genet. 2:e177 10.1371/journal.pgen.0020177 - DOI - PMC - PubMed
-
- Balciunas D., Wangensteen K.J., Wilber A., Bell J., Geurts A., Sivasubbu S., Wang X., Hackett P.B., Largaespada D.A., McIvor R.S., Ekker S.C. 2006. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2:e169 10.1371/journal.pgen.0020169 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

