Common gene rearrangements in prostate cancer
- PMID: 21859993
- PMCID: PMC4874145
- DOI: 10.1200/JCO.2011.35.1916
Common gene rearrangements in prostate cancer
Abstract
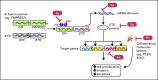
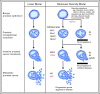
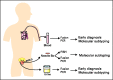
Prostate cancer is a common heterogeneous disease, and most patients diagnosed in the post prostate-specific antigen (PSA) era present with clinically localized disease, the majority of which do well regardless of treatment regimen undertaken. Overall, those with advanced prostate cancer at time of diagnosis do poorly after androgen withdrawal therapy. Understanding the biologic underpinning of prostate cancer is necessary to best determine the risk of disease progression and would be advantageous for the development of novel therapeutic approaches to impede or prevent disease. This review focuses on the recently identified common ETS and non-ETS gene rearrangements in prostate cancer. Although multiple molecular alterations have been detected in prostate cancer, a detailed understanding of gene fusion prostate cancer should help explain the clinical and biologic diversity, providing a rationale for a molecular subclassification of the disease.
Conflict of interest statement
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Figures



References
-
- Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. - PubMed
-
- Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. - PubMed
-
- Rubin MA, Chinnaiyan AM. Bioinformatics approach leads to the discovery of the TMPRSS2:ETS gene fusion in prostate cancer. Lab Invest. 2006;86:1099–1102. - PubMed
-
- Hanauer DA, Rhodes DR, Sinha-Kumar C, et al. Bioinformatics approaches in the study of cancer. Curr Mol Med. 2007;7:133–141. - PubMed
-
- Tomlins SA, Mehra R, Rhodes DR, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

