The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation
- PMID: 21860393
- PMCID: PMC3545438
- DOI: 10.1038/nrm3173
The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation
Erratum in
- Nat Rev Mol Cell Biol. 2011 Oct;12(10):686
Abstract
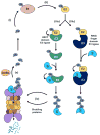
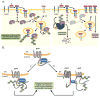
Ubiquitylation (also known as ubiquitination) regulates essentially all of the intracellular processes in eukaryotes through highly specific modification of numerous cellular proteins, which is often tightly regulated in a spatial and temporal manner. Although most often associated with proteasomal degradation, ubiquitylation frequently serves non-proteolytic functions. In light of its central roles in cellular regulation, it has not been surprising to find that many of the components of the ubiquitin system itself are regulated by ubiquitylation. This observation has broad implications for pathophysiology.
Figures
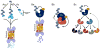


References
-
- Shabek N, Iwai K, Ciechanover A. Ubiquitin is degraded by the ubiquitin system as a monomer and as part of its conjugated target. Biochem Biophys Res Commun. 2007;363:425–431. - PubMed
-
- Hershko A, Eytan E, Ciechanover A, Haas AL. Immunochemical analysis of the turnover of ubiquitin-protein conjugates in intact cells. Relationship to the breakdown of abnormal proteins. J Biol Chem. 1982;257:13964–13970. - PubMed
-
The first description of the role of the ubiquitin proteolytic system in the degradation of proteins in nucleated cells. All prior studies describing the roles of the system were carried out using reticulocytes - the terminally differentiating red blood cell.
-
- Haas AL, Bright PM. The dynamics of ubiquitin pools within cultured human lung fibroblasts. J Biol Chem. 1987;262:345–351. - PubMed
-
- Patel MB, Majetschak M. Distribution and interrelationship of ubiquitin proteasome pathway component activities and ubiquitin pools in various porcine tissues. Physiol Res. 2007;56:341–350. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

