HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs
- PMID: 21860410
- PMCID: PMC3223555
- DOI: 10.1038/onc.2011.365
HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs
Erratum in
-
Correction: HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs.Oncogene. 2021 Feb;40(8):1552-1553. doi: 10.1038/s41388-020-01618-z. Oncogene. 2021. PMID: 33452464 No abstract available.
Retraction in
-
Retraction Note: HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs.Oncogene. 2023 Jul;42(28):2234. doi: 10.1038/s41388-023-02720-8. Oncogene. 2023. PMID: 37221224 Free PMC article. No abstract available.
Abstract
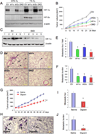
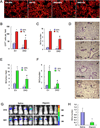
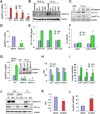
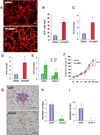
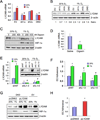
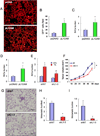
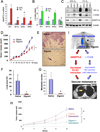
Most cases of breast cancer (BrCa) mortality are due to vascular metastasis. BrCa cells must intravasate through endothelial cells (ECs) to enter a blood vessel in the primary tumor and then adhere to ECs and extravasate at the metastatic site. In this study we demonstrate that inhibition of hypoxia-inducible factor (HIF) activity in BrCa cells by RNA interference or digoxin treatment inhibits primary tumor growth and also inhibits the metastasis of BrCa cells to the lungs by blocking the expression of angiopoietin-like 4 (ANGPTL4) and L1 cell adhesion molecule (L1CAM). ANGPTL4 is a secreted factor that inhibits EC-EC interaction, whereas L1CAM increases the adherence of BrCa cells to ECs. Interference with HIF, ANGPTL4 or L1CAM expression inhibits vascular metastasis of BrCa cells to the lungs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Modified Bu-Fei decoction inhibits lung metastasis via suppressing angiopoietin-like 4.Phytomedicine. 2022 Nov;106:154409. doi: 10.1016/j.phymed.2022.154409. Epub 2022 Aug 28. Phytomedicine. 2022. PMID: 36070661
-
Hypoxia-Inducible Factor-Dependent Expression of Angiopoietin-Like 4 by Conjunctival Epithelial Cells Promotes the Angiogenic Phenotype of Pterygia.Invest Ophthalmol Vis Sci. 2017 Sep 1;58(11):4514-4523. doi: 10.1167/iovs.17-21974. Invest Ophthalmol Vis Sci. 2017. PMID: 28873177 Free PMC article.
-
Inhibitors of hypoxia-inducible factor 1 block breast cancer metastatic niche formation and lung metastasis.J Mol Med (Berl). 2012 Jul;90(7):803-15. doi: 10.1007/s00109-011-0855-y. Epub 2012 Jan 10. J Mol Med (Berl). 2012. PMID: 22231744 Free PMC article.
-
Molecular mechanisms mediating metastasis of hypoxic breast cancer cells.Trends Mol Med. 2012 Sep;18(9):534-43. doi: 10.1016/j.molmed.2012.08.001. Epub 2012 Aug 23. Trends Mol Med. 2012. PMID: 22921864 Free PMC article. Review.
-
Hypoxia-inducible factor 1 and breast cancer metastasis.J Zhejiang Univ Sci B. 2015 Jan;16(1):32-43. doi: 10.1631/jzus.B1400221. J Zhejiang Univ Sci B. 2015. PMID: 25559953 Free PMC article. Review.
Cited by
-
Danlou Tablet Improves Chronic Intermittent Hypoxia-Induced Dyslipidemia and Arteriosclerosis by HIF-1α-Angptl4 mRNA Signaling Pathway.Chin J Integr Med. 2022 Jun;28(6):509-517. doi: 10.1007/s11655-020-3255-8. Epub 2020 Jul 4. Chin J Integr Med. 2022. PMID: 32623702
-
Role of L1 cell adhesion molecule (L1CAM) in the metastatic cascade: promotion of dissemination, colonization, and metastatic growth.Clin Exp Metastasis. 2014 Jan;31(1):87-100. doi: 10.1007/s10585-013-9613-6. Epub 2013 Sep 4. Clin Exp Metastasis. 2014. PMID: 24002299
-
Chemotherapy induces enrichment of CD47+/CD73+/PDL1+ immune evasive triple-negative breast cancer cells.Proc Natl Acad Sci U S A. 2018 Feb 6;115(6):E1239-E1248. doi: 10.1073/pnas.1718197115. Epub 2018 Jan 24. Proc Natl Acad Sci U S A. 2018. PMID: 29367423 Free PMC article.
-
Evaluating the cancer therapeutic potential of cardiac glycosides.Biomed Res Int. 2014;2014:794930. doi: 10.1155/2014/794930. Epub 2014 May 8. Biomed Res Int. 2014. PMID: 24895612 Free PMC article. Review.
-
Angiopoietin-like 4 promotes angiogenesis in the tendon and is increased in cyclically loaded tendon fibroblasts.J Physiol. 2016 Jun 1;594(11):2971-83. doi: 10.1113/JP271752. Epub 2016 Jan 18. J Physiol. 2016. PMID: 26670924 Free PMC article.
References
-
- Belanger AJ, Lu H, Date T, Liu LX, Vincent KA, Akita GY, et al. Hypoxia up-regulates expression of peroxisome proliferator-activated receptor gamma angiopoietin-related gene (PGAR) in cardiomyocytes: role of hypoxia inducible factor 1α. J Mol Cell Cardiol. 2002;34:765–774. - PubMed
-
- Bos R, van der Groep P, Greijer AE, Shvarts A, Meijer S, Pinedo HM, et al. Levels of hypoxia-inducible factor 1α independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer. 2003;97:1573–1581. - PubMed
-
- Bos R, Zhong H, Hanrahan CF, Mommers EC, Semenza GL, Pinedo HM, et al. Levels of hypoxia-inducible factor 1α during breast carcinogenesis. J Natl Cancer Inst. 2001;93:309–314. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

