FOXO3a represses VEGF expression through FOXM1-dependent and -independent mechanisms in breast cancer
- PMID: 21860419
- PMCID: PMC3232453
- DOI: 10.1038/onc.2011.368
FOXO3a represses VEGF expression through FOXM1-dependent and -independent mechanisms in breast cancer
Erratum in
-
Correction: FOXO3a represses VEGF expression through FOXM1-dependent and -independent mechanisms in breast cancer.Oncogene. 2019 Jun;38(25):5111-5112. doi: 10.1038/s41388-019-0770-1. Oncogene. 2019. PMID: 30867566
Abstract
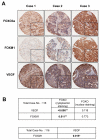
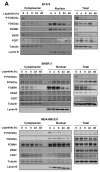
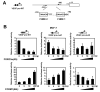
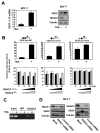
Vascular endothelial growth factor (VEGF) has a central role in breast cancer development and progression, but the mechanisms that control its expression are poorly understood. Breast cancer tissue microarrays revealed an inverse correlation between the Forkhead transcription factor Forkhead box class O (FOXO)3a and VEGF expression. Using the lapatinib-sensitive breast cancer cell lines BT474 and SKBR3 as model systems, we tested the possibility that VEGF expression is negatively regulated by FOXO3a. Lapatinib treatment of BT474 or SKBR3 cells resulted in nuclear translocation and activation of FOXO3a, followed by a reduction in VEGF expression. Transient transfection and inducible expression experiments showed that FOXO3a represses the proximal VEGF promoter, whereas another Forkhead member, FOXM1, induces VEGF expression. Chromatin immunoprecipitation and oligonucleotide pull-down assays showed that both FOXO3a and FOXM1 bind a consensus Forkhead response element (FHRE) in the VEGF promoter. Upon lapatinib stimulation, activated FOXO3a displaces FOXM1 bound to the FHRE before recruiting histone deacetylase 2 (HDAC2) to the promoter, leading to decreased histones H3 and H4 acetylation, and concomitant transcriptional inhibition of VEGF. These results show that FOXO3a-dependent repression of target genes in breast cancer cells, such as VEGF, involves competitive displacement of DNA-bound FOXM1 and active recruitment of transcriptional repressor complexes.
Figures





Similar articles
-
Gefitinib (Iressa) represses FOXM1 expression via FOXO3a in breast cancer.Mol Cancer Ther. 2009 Mar;8(3):582-91. doi: 10.1158/1535-7163.MCT-08-0805. Epub 2009 Mar 10. Mol Cancer Ther. 2009. PMID: 19276163
-
The Forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells.J Biol Chem. 2006 Sep 1;281(35):25167-76. doi: 10.1074/jbc.M603906200. Epub 2006 Jun 28. J Biol Chem. 2006. PMID: 16809346
-
[Effects of AG1478 on the expression of FOXM1 gene via FOXO3a in non-small cell lung cancer cells].Zhonghua Zhong Liu Za Zhi. 2013 Aug;35(8):572-8. Zhonghua Zhong Liu Za Zhi. 2013. PMID: 24314213 Chinese.
-
Insights into a Critical Role of the FOXO3a-FOXM1 Axis in DNA Damage Response and Genotoxic Drug Resistance.Curr Drug Targets. 2016;17(2):164-77. doi: 10.2174/1389450115666141122211549. Curr Drug Targets. 2016. PMID: 25418858 Free PMC article. Review.
-
Role and regulation of the forkhead transcription factors FOXO3a and FOXM1 in carcinogenesis and drug resistance.Chin J Cancer. 2013 Jul;32(7):365-70. doi: 10.5732/cjc.012.10277. Epub 2013 May 27. Chin J Cancer. 2013. PMID: 23706767 Free PMC article. Review.
Cited by
-
Chasing the FOXO3: Insights into Its New Mitochondrial Lair in Colorectal Cancer Landscape.Cancers (Basel). 2019 Mar 23;11(3):414. doi: 10.3390/cancers11030414. Cancers (Basel). 2019. PMID: 30909600 Free PMC article. Review.
-
Genetics of breast cancer bone metastasis: a sequential multistep pattern.Clin Exp Metastasis. 2014 Jun;31(5):595-612. doi: 10.1007/s10585-014-9642-9. Epub 2014 Feb 4. Clin Exp Metastasis. 2014. PMID: 24493024 Review.
-
The link between FOXJ1 expression level in bladder carcinoma and tumor recurrence.Oncol Lett. 2018 Feb;15(2):1483-1486. doi: 10.3892/ol.2017.7504. Epub 2017 Nov 29. Oncol Lett. 2018. Retraction in: Oncol Lett. 2018 Aug;16(2):2764. doi: 10.3892/ol.2018.8920. PMID: 29434839 Free PMC article. Retracted.
-
Effect of high-intensity exercise on aged mouse brain mitochondria, neurogenesis, and inflammation.Neurobiol Aging. 2014 Nov;35(11):2574-2583. doi: 10.1016/j.neurobiolaging.2014.05.033. Epub 2014 Jun 10. Neurobiol Aging. 2014. PMID: 25002036 Free PMC article.
-
Forkhead box proteins: tuning forks for transcriptional harmony.Nat Rev Cancer. 2013 Jul;13(7):482-95. doi: 10.1038/nrc3539. Nat Rev Cancer. 2013. PMID: 23792361 Review.
References
-
- Arden KC. FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene. 2008;27:2345–2350. - PubMed
-
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. - PubMed
-
- Burgering BM. A brief introduction to FOXOlogy. Oncogene. 2008;27:2258–2262. - PubMed
-
- Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. - PubMed
-
- Ciardiello F. Epidermal growth factor receptor inhibitors in cancer treatment. Future Oncol. 2005;1:221–234. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

