Simulation of DNA Extension in Nanochannels
- PMID: 21860535
- PMCID: PMC3158571
- DOI: 10.1021/ma201277e
Simulation of DNA Extension in Nanochannels
Abstract
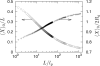
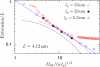
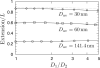
We have used a realistic model for double stranded DNA and Monte Carlo simulations to compute the extension (mean span) of a DNA molecule confined in a nanochannel over the full range of confinement in a high ionic strength buffer. The simulation data for square nanochannels resolve the apparent contradiction between prior simulation studies and the predictions from Flory theory, demonstrating the existence of two transition regimes between weak confinement (the de Gennes regime) and strong confinement (the Odijk regime). The simulation data for rectangular nanochannels support the use of the geometric mean for mapping data obtained in rectangular channels onto models developed for cylinders. The comparison of our results with experimental data illuminates the challenges in applying models for confined, neutral polymers to polyelectrolytes. Using a Flory-type approach, we also provide an improved scaling result for the relaxation time in the transition regime close to that found in experiments.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
