The ACR11 encodes a novel type of chloroplastic ACT domain repeat protein that is coordinately expressed with GLN2 in Arabidopsis
- PMID: 21861936
- PMCID: PMC3173338
- DOI: 10.1186/1471-2229-11-118
The ACR11 encodes a novel type of chloroplastic ACT domain repeat protein that is coordinately expressed with GLN2 in Arabidopsis
Abstract
Background: The ACT domain, named after bacterial aspartate kinase, chorismate mutase and TyrA (prephenate dehydrogenase), is a regulatory domain that serves as an amino acid-binding site in feedback-regulated amino acid metabolic enzymes. We have previously identified a novel type of ACT domain-containing protein family, the ACT domain repeat (ACR) protein family, in Arabidopsis. Members of the ACR family, ACR1 to ACR8, contain four copies of the ACT domain that extend throughout the entire polypeptide. Here, we describe the identification of four novel ACT domain-containing proteins, namely ACR9 to ACR12, in Arabidopsis. The ACR9 and ACR10 proteins contain three copies of the ACT domain, whereas the ACR11 and ACR12 proteins have a putative transit peptide followed by two copies of the ACT domain. The functions of these plant ACR proteins are largely unknown.
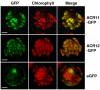
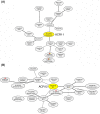
Results: The ACR11 and ACR12 proteins are predicted to target to chloroplasts. We used protoplast transient expression assay to demonstrate that the Arabidopsis ACR11- and ACR12-green fluorescent fusion proteins are localized to the chloroplast. Analysis of an ACR11 promoter-β-glucuronidase (GUS) fusion in transgenic Arabidopsis revealed that the GUS activity was mainly detected in mature leaves and sepals. Interestingly, coexpression analysis revealed that the GLN2, which encodes a chloroplastic glutamine synthetase, has the highest mutual rank in the coexpressed gene network connected to ACR11. We used RNA gel blot analysis to confirm that the expression pattern of ACR11 is similar to that of GLN2 in various organs from 6-week-old Arabidopsis. Moreover, the expression of ACR11 and GLN2 is highly co-regulated by sucrose and light/dark treatments in 2-week-old Arabidopsis seedlings.
Conclusions: This study reports the identification of four novel ACT domain repeat proteins, ACR9 to ACR12, in Arabidopsis. The ACR11 and ACR12 proteins are localized to the chloroplast, and the expression of ACR11 and GLN2 is highly coordinated. These results suggest that the ACR11 and GLN2 genes may belong to the same functional module. The Arabidopsis ACR11 protein may function as a regulatory protein that is related to glutamine metabolism or signaling in the chloroplast.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

