Visualizing kinetochore architecture
- PMID: 21862320
- PMCID: PMC3189262
- DOI: 10.1016/j.sbi.2011.07.009
Visualizing kinetochore architecture
Abstract
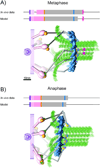
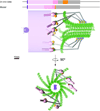
Kinetochores are large macromolecular assemblies that link chromosomes to spindle microtubules (MTs) during mitosis. Here we review recent advances in the study of core MT-binding kinetochore complexes using electron microcopy methods in vitro and nanometer-accuracy fluorescence microscopy in vivo. We synthesize these findings in novel three-dimensional models of both the budding yeast and vertebrate kinetochore in different stages of mitosis. There is a growing consensus that kinetochores are highly dynamic, supra-molecular machines that undergo dramatic structural rearrangements in response to MT capture and spindle forces during mitosis.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

References
-
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. - PubMed
-
- Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. - PubMed
-
- Koshland DE, Mitchison TJ, Kirschner MW. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

