Snf2-family proteins: chromatin remodellers for any occasion
- PMID: 21862382
- PMCID: PMC4162295
- DOI: 10.1016/j.cbpa.2011.07.022
Snf2-family proteins: chromatin remodellers for any occasion
Abstract
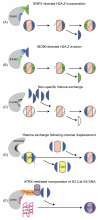
Chromatin facilitates the housing of eukaryotic DNA within the nucleus and restricts access to the underlying sequences. Thus, the regulation of chromatin structure provides an excellent platform for regulating processes that require information stored within genomic DNA. Snf2 proteins are a family of helicase-like proteins that direct energy derived from ATP hydrolysis into the mechanical remodelling of chromatin structure. Here, we highlight some of the recent discoveries regarding this family of proteins and show Snf2 proteins have roles in many aspects of genetic metabolism. Recent developments include new insights into the mechanism for nucleosome spacing and histone dimer exchange; together with growing evidence for the involvement of Snf2 proteins in DNA repair.
Copyright © 2011. Published by Elsevier Ltd.
Figures

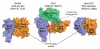
References
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. - PubMed
-
- Justin N, De Marco V, Aasland R, Gamblin SJ. Reading, writing and editing methylated lysines on histone tails: new insights from recent structural studies. Current opinion in structural biology. 2010;20:730–738. - PubMed
-
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature reviews. Genetics. 2008;9:465–476. - PubMed
-
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature reviews. Genetics. 2011;12:7–18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

